
Banna virus (strain Indonesia/JKT-6423/1980) (BAV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Seadornavirus; Banna virus
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins
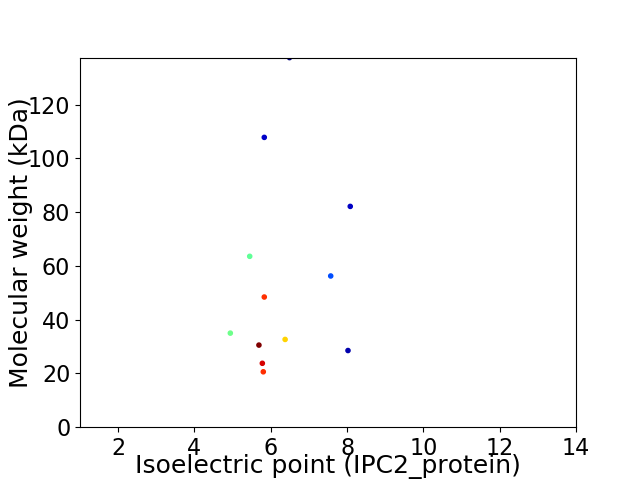
Virtual 2D-PAGE plot for 12 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
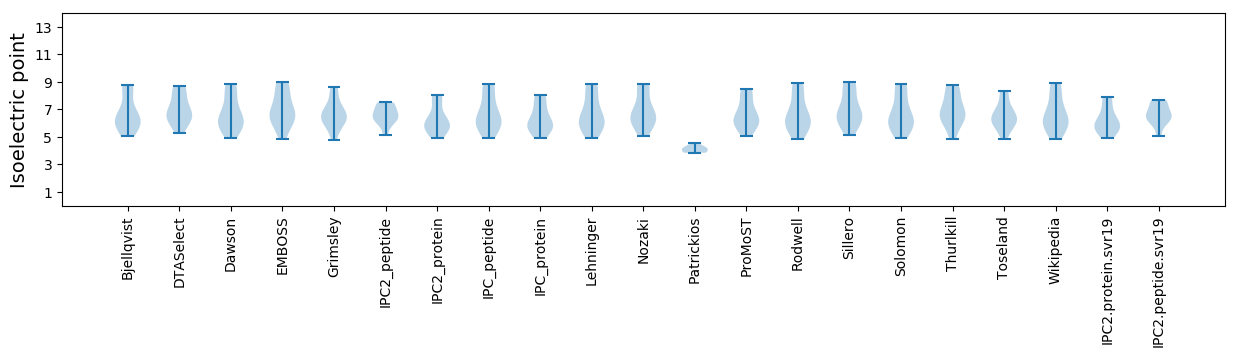
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q9YWQ1|SPIKE_BAVJK Spike protein VP9 OS=Banna virus (strain Indonesia/JKT-6423/1980) OX=649604 GN=Segment-9 PE=4 SV=1
MM1 pKa = 7.58NNGQATITRR10 pKa = 11.84NGGRR14 pKa = 11.84FEE16 pKa = 3.86IRR18 pKa = 11.84CRR20 pKa = 11.84HH21 pKa = 6.29LDD23 pKa = 3.27RR24 pKa = 11.84DD25 pKa = 3.85YY26 pKa = 11.97TMPLPNATSNDD37 pKa = 3.34NFLDD41 pKa = 4.43CIKK44 pKa = 10.76FITEE48 pKa = 4.25CVGFDD53 pKa = 3.58YY54 pKa = 11.12VSSGFKK60 pKa = 10.73LIANVNDD67 pKa = 3.91FQHH70 pKa = 6.83LNGNSTLLIGKK81 pKa = 6.81TKK83 pKa = 10.22IGPLILKK90 pKa = 9.61KK91 pKa = 10.56VRR93 pKa = 11.84SLPCCNDD100 pKa = 2.73ALFRR104 pKa = 11.84NEE106 pKa = 3.65FRR108 pKa = 11.84ILAKK112 pKa = 9.4MHH114 pKa = 6.73GILRR118 pKa = 11.84LKK120 pKa = 10.76NDD122 pKa = 3.79VNGHH126 pKa = 5.98KK127 pKa = 10.52YY128 pKa = 10.62GIILEE133 pKa = 4.24RR134 pKa = 11.84CYY136 pKa = 10.65KK137 pKa = 10.4PKK139 pKa = 10.47INFSNFVTAINDD151 pKa = 3.43LDD153 pKa = 3.94VFHH156 pKa = 7.5SSNQHH161 pKa = 6.12LLHH164 pKa = 7.23GDD166 pKa = 3.77ANPDD170 pKa = 3.48NIMSDD175 pKa = 3.34SEE177 pKa = 5.06GYY179 pKa = 10.43LKK181 pKa = 10.89LVDD184 pKa = 4.09PVCLLEE190 pKa = 4.24NQVNMVNIEE199 pKa = 4.14YY200 pKa = 10.47EE201 pKa = 4.31SLTQEE206 pKa = 3.92AEE208 pKa = 3.81KK209 pKa = 10.78KK210 pKa = 10.59VFINSLLQLVEE221 pKa = 4.27KK222 pKa = 10.74QMSATIDD229 pKa = 3.64EE230 pKa = 4.82IYY232 pKa = 11.44VNLKK236 pKa = 9.35EE237 pKa = 4.33VNPSFNLEE245 pKa = 4.18HH246 pKa = 6.51GLKK249 pKa = 10.75LSDD252 pKa = 4.19LLDD255 pKa = 4.56NIDD258 pKa = 4.9VYY260 pKa = 11.67NSDD263 pKa = 3.01HH264 pKa = 6.62WKK266 pKa = 10.84LMLNHH271 pKa = 6.77RR272 pKa = 11.84PMMPEE277 pKa = 4.24LSVLNDD283 pKa = 3.14LTYY286 pKa = 11.04YY287 pKa = 9.85DD288 pKa = 3.74TGEE291 pKa = 4.02VRR293 pKa = 11.84DD294 pKa = 4.92LVTEE298 pKa = 4.48DD299 pKa = 5.56LDD301 pKa = 6.27DD302 pKa = 4.69EE303 pKa = 5.88DD304 pKa = 5.88DD305 pKa = 3.7VV306 pKa = 4.58
MM1 pKa = 7.58NNGQATITRR10 pKa = 11.84NGGRR14 pKa = 11.84FEE16 pKa = 3.86IRR18 pKa = 11.84CRR20 pKa = 11.84HH21 pKa = 6.29LDD23 pKa = 3.27RR24 pKa = 11.84DD25 pKa = 3.85YY26 pKa = 11.97TMPLPNATSNDD37 pKa = 3.34NFLDD41 pKa = 4.43CIKK44 pKa = 10.76FITEE48 pKa = 4.25CVGFDD53 pKa = 3.58YY54 pKa = 11.12VSSGFKK60 pKa = 10.73LIANVNDD67 pKa = 3.91FQHH70 pKa = 6.83LNGNSTLLIGKK81 pKa = 6.81TKK83 pKa = 10.22IGPLILKK90 pKa = 9.61KK91 pKa = 10.56VRR93 pKa = 11.84SLPCCNDD100 pKa = 2.73ALFRR104 pKa = 11.84NEE106 pKa = 3.65FRR108 pKa = 11.84ILAKK112 pKa = 9.4MHH114 pKa = 6.73GILRR118 pKa = 11.84LKK120 pKa = 10.76NDD122 pKa = 3.79VNGHH126 pKa = 5.98KK127 pKa = 10.52YY128 pKa = 10.62GIILEE133 pKa = 4.24RR134 pKa = 11.84CYY136 pKa = 10.65KK137 pKa = 10.4PKK139 pKa = 10.47INFSNFVTAINDD151 pKa = 3.43LDD153 pKa = 3.94VFHH156 pKa = 7.5SSNQHH161 pKa = 6.12LLHH164 pKa = 7.23GDD166 pKa = 3.77ANPDD170 pKa = 3.48NIMSDD175 pKa = 3.34SEE177 pKa = 5.06GYY179 pKa = 10.43LKK181 pKa = 10.89LVDD184 pKa = 4.09PVCLLEE190 pKa = 4.24NQVNMVNIEE199 pKa = 4.14YY200 pKa = 10.47EE201 pKa = 4.31SLTQEE206 pKa = 3.92AEE208 pKa = 3.81KK209 pKa = 10.78KK210 pKa = 10.59VFINSLLQLVEE221 pKa = 4.27KK222 pKa = 10.74QMSATIDD229 pKa = 3.64EE230 pKa = 4.82IYY232 pKa = 11.44VNLKK236 pKa = 9.35EE237 pKa = 4.33VNPSFNLEE245 pKa = 4.18HH246 pKa = 6.51GLKK249 pKa = 10.75LSDD252 pKa = 4.19LLDD255 pKa = 4.56NIDD258 pKa = 4.9VYY260 pKa = 11.67NSDD263 pKa = 3.01HH264 pKa = 6.62WKK266 pKa = 10.84LMLNHH271 pKa = 6.77RR272 pKa = 11.84PMMPEE277 pKa = 4.24LSVLNDD283 pKa = 3.14LTYY286 pKa = 11.04YY287 pKa = 9.85DD288 pKa = 3.74TGEE291 pKa = 4.02VRR293 pKa = 11.84DD294 pKa = 4.92LVTEE298 pKa = 4.48DD299 pKa = 5.56LDD301 pKa = 6.27DD302 pKa = 4.69EE303 pKa = 5.88DD304 pKa = 5.88DD305 pKa = 3.7VV306 pKa = 4.58
Molecular weight: 34.99 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q9INI5|VP2_BAVJK Major inner capsid protein VP2 OS=Banna virus (strain Indonesia/JKT-6423/1980) OX=649604 GN=Segment-2 PE=4 SV=1
MM1 pKa = 7.61EE2 pKa = 5.84LFSDD6 pKa = 3.55SGSIVEE12 pKa = 4.36NFKK15 pKa = 11.04EE16 pKa = 4.34RR17 pKa = 11.84INKK20 pKa = 9.24LVFDD24 pKa = 4.1YY25 pKa = 11.16SLNHH29 pKa = 5.68GGFRR33 pKa = 11.84KK34 pKa = 8.52TYY36 pKa = 9.17KK37 pKa = 9.19IQRR40 pKa = 11.84DD41 pKa = 3.27RR42 pKa = 11.84VYY44 pKa = 11.67YY45 pKa = 9.25MLGDD49 pKa = 3.5AHH51 pKa = 6.7HH52 pKa = 7.28ANLSGKK58 pKa = 9.98CLMLYY63 pKa = 10.4NSEE66 pKa = 4.0KK67 pKa = 10.9DD68 pKa = 3.02IFEE71 pKa = 4.24GLGFKK76 pKa = 10.71VKK78 pKa = 10.3GSRR81 pKa = 11.84INVSKK86 pKa = 9.03TQILRR91 pKa = 11.84NYY93 pKa = 8.43EE94 pKa = 3.98INFEE98 pKa = 4.39TIIGVEE104 pKa = 3.99PNGLKK109 pKa = 9.92TISTAKK115 pKa = 10.14DD116 pKa = 3.16VKK118 pKa = 10.45KK119 pKa = 10.81LYY121 pKa = 10.41DD122 pKa = 3.18IYY124 pKa = 11.03SYY126 pKa = 11.08KK127 pKa = 10.67SSLHH131 pKa = 6.18PFDD134 pKa = 6.02DD135 pKa = 5.81FMAHH139 pKa = 7.35CINRR143 pKa = 11.84WGMSIPASLEE153 pKa = 3.85RR154 pKa = 11.84IIKK157 pKa = 10.12SEE159 pKa = 3.96IIKK162 pKa = 10.07VRR164 pKa = 11.84SGVLNRR170 pKa = 11.84NSEE173 pKa = 4.51LYY175 pKa = 10.73NYY177 pKa = 9.61IPTVDD182 pKa = 3.5ASFSEE187 pKa = 4.79MSRR190 pKa = 11.84GPANVILTDD199 pKa = 3.79GKK201 pKa = 10.65LVPDD205 pKa = 4.64GTCFGPILSKK215 pKa = 10.81SVEE218 pKa = 4.06DD219 pKa = 3.71PRR221 pKa = 11.84LKK223 pKa = 11.05NEE225 pKa = 3.86FRR227 pKa = 11.84SKK229 pKa = 10.8GLIMVHH235 pKa = 7.33DD236 pKa = 4.18YY237 pKa = 10.97FILIGEE243 pKa = 4.55SPGPHH248 pKa = 5.03YY249 pKa = 10.78KK250 pKa = 10.46KK251 pKa = 8.89YY252 pKa = 9.71TKK254 pKa = 7.8MTKK257 pKa = 10.43DD258 pKa = 3.11NTIFWDD264 pKa = 3.96PRR266 pKa = 11.84RR267 pKa = 11.84TDD269 pKa = 3.04HH270 pKa = 7.02KK271 pKa = 10.45FNNVVSYY278 pKa = 9.21FKK280 pKa = 10.84KK281 pKa = 9.98EE282 pKa = 4.01NIRR285 pKa = 11.84DD286 pKa = 3.79VVEE289 pKa = 4.17YY290 pKa = 8.22TTDD293 pKa = 3.1ALNRR297 pKa = 11.84GLKK300 pKa = 9.91PLVLIDD306 pKa = 3.62IRR308 pKa = 11.84KK309 pKa = 9.96DD310 pKa = 3.09KK311 pKa = 10.7PKK313 pKa = 10.9NLNTPEE319 pKa = 4.16GAIEE323 pKa = 4.15WEE325 pKa = 4.36RR326 pKa = 11.84MVHH329 pKa = 7.07DD330 pKa = 5.4DD331 pKa = 3.76NNLIIDD337 pKa = 4.22MVNALDD343 pKa = 4.89KK344 pKa = 10.97RR345 pKa = 11.84VTVCAKK351 pKa = 10.06LRR353 pKa = 11.84PAFMQVGSMRR363 pKa = 11.84KK364 pKa = 8.67LLRR367 pKa = 11.84PVRR370 pKa = 11.84ILPLPYY376 pKa = 9.98LRR378 pKa = 11.84RR379 pKa = 11.84STAEE383 pKa = 3.44FNMFVPNEE391 pKa = 3.6ALMNGNEE398 pKa = 4.45IYY400 pKa = 10.7DD401 pKa = 3.58VTYY404 pKa = 11.16DD405 pKa = 3.53DD406 pKa = 4.63LVRR409 pKa = 11.84MSSEE413 pKa = 3.93VFVLKK418 pKa = 10.86NIIGGLYY425 pKa = 10.64NMYY428 pKa = 10.47LKK430 pKa = 10.85DD431 pKa = 3.22MHH433 pKa = 7.11LNLGVVNKK441 pKa = 10.26SVSLSDD447 pKa = 3.37GSSAIWSLSNINNEE461 pKa = 4.25RR462 pKa = 11.84ISNFNFNNFLYY473 pKa = 10.33AAPYY477 pKa = 10.6SDD479 pKa = 4.27FATSSVKK486 pKa = 10.39RR487 pKa = 11.84HH488 pKa = 4.98FKK490 pKa = 9.63GRR492 pKa = 11.84NYY494 pKa = 10.74SDD496 pKa = 2.98WCLNILDD503 pKa = 4.57EE504 pKa = 4.76VNLKK508 pKa = 10.88DD509 pKa = 4.12GVYY512 pKa = 10.25LVPLYY517 pKa = 10.79AIVGGGQITSHH528 pKa = 6.86DD529 pKa = 4.03FVNAIITDD537 pKa = 3.74QEE539 pKa = 3.96QLIDD543 pKa = 3.71FTQSEE548 pKa = 4.86RR549 pKa = 11.84ALSTQVVKK557 pKa = 10.75LVSFILKK564 pKa = 10.48DD565 pKa = 3.34SFTAKK570 pKa = 9.98GLNWTEE576 pKa = 3.29IDD578 pKa = 3.35NEE580 pKa = 3.79IRR582 pKa = 11.84NRR584 pKa = 11.84RR585 pKa = 11.84LSSLSGVGFTVTKK598 pKa = 10.1MLDD601 pKa = 3.34GKK603 pKa = 10.59VLVDD607 pKa = 3.66GKK609 pKa = 10.72VVTVSGHH616 pKa = 4.53MLYY619 pKa = 10.1ILLGSILGLPYY630 pKa = 10.6GIKK633 pKa = 10.13KK634 pKa = 9.14YY635 pKa = 10.78LKK637 pKa = 9.99EE638 pKa = 3.99IEE640 pKa = 4.44LNILKK645 pKa = 10.07PGSSYY650 pKa = 10.95EE651 pKa = 3.93RR652 pKa = 11.84GVGGRR657 pKa = 11.84VWHH660 pKa = 6.14GLISHH665 pKa = 6.58YY666 pKa = 10.79LAVDD670 pKa = 3.59CVIDD674 pKa = 4.55VIDD677 pKa = 4.4EE678 pKa = 4.4YY679 pKa = 10.7MVCTYY684 pKa = 10.44EE685 pKa = 5.62DD686 pKa = 3.62RR687 pKa = 11.84SKK689 pKa = 11.58LNVVLRR695 pKa = 11.84YY696 pKa = 9.79VKK698 pKa = 10.64SKK700 pKa = 10.72LLEE703 pKa = 4.63LGSKK707 pKa = 9.79YY708 pKa = 10.61DD709 pKa = 3.28VYY711 pKa = 11.43LSVDD715 pKa = 3.18EE716 pKa = 4.78RR717 pKa = 11.84LVLL720 pKa = 4.14
MM1 pKa = 7.61EE2 pKa = 5.84LFSDD6 pKa = 3.55SGSIVEE12 pKa = 4.36NFKK15 pKa = 11.04EE16 pKa = 4.34RR17 pKa = 11.84INKK20 pKa = 9.24LVFDD24 pKa = 4.1YY25 pKa = 11.16SLNHH29 pKa = 5.68GGFRR33 pKa = 11.84KK34 pKa = 8.52TYY36 pKa = 9.17KK37 pKa = 9.19IQRR40 pKa = 11.84DD41 pKa = 3.27RR42 pKa = 11.84VYY44 pKa = 11.67YY45 pKa = 9.25MLGDD49 pKa = 3.5AHH51 pKa = 6.7HH52 pKa = 7.28ANLSGKK58 pKa = 9.98CLMLYY63 pKa = 10.4NSEE66 pKa = 4.0KK67 pKa = 10.9DD68 pKa = 3.02IFEE71 pKa = 4.24GLGFKK76 pKa = 10.71VKK78 pKa = 10.3GSRR81 pKa = 11.84INVSKK86 pKa = 9.03TQILRR91 pKa = 11.84NYY93 pKa = 8.43EE94 pKa = 3.98INFEE98 pKa = 4.39TIIGVEE104 pKa = 3.99PNGLKK109 pKa = 9.92TISTAKK115 pKa = 10.14DD116 pKa = 3.16VKK118 pKa = 10.45KK119 pKa = 10.81LYY121 pKa = 10.41DD122 pKa = 3.18IYY124 pKa = 11.03SYY126 pKa = 11.08KK127 pKa = 10.67SSLHH131 pKa = 6.18PFDD134 pKa = 6.02DD135 pKa = 5.81FMAHH139 pKa = 7.35CINRR143 pKa = 11.84WGMSIPASLEE153 pKa = 3.85RR154 pKa = 11.84IIKK157 pKa = 10.12SEE159 pKa = 3.96IIKK162 pKa = 10.07VRR164 pKa = 11.84SGVLNRR170 pKa = 11.84NSEE173 pKa = 4.51LYY175 pKa = 10.73NYY177 pKa = 9.61IPTVDD182 pKa = 3.5ASFSEE187 pKa = 4.79MSRR190 pKa = 11.84GPANVILTDD199 pKa = 3.79GKK201 pKa = 10.65LVPDD205 pKa = 4.64GTCFGPILSKK215 pKa = 10.81SVEE218 pKa = 4.06DD219 pKa = 3.71PRR221 pKa = 11.84LKK223 pKa = 11.05NEE225 pKa = 3.86FRR227 pKa = 11.84SKK229 pKa = 10.8GLIMVHH235 pKa = 7.33DD236 pKa = 4.18YY237 pKa = 10.97FILIGEE243 pKa = 4.55SPGPHH248 pKa = 5.03YY249 pKa = 10.78KK250 pKa = 10.46KK251 pKa = 8.89YY252 pKa = 9.71TKK254 pKa = 7.8MTKK257 pKa = 10.43DD258 pKa = 3.11NTIFWDD264 pKa = 3.96PRR266 pKa = 11.84RR267 pKa = 11.84TDD269 pKa = 3.04HH270 pKa = 7.02KK271 pKa = 10.45FNNVVSYY278 pKa = 9.21FKK280 pKa = 10.84KK281 pKa = 9.98EE282 pKa = 4.01NIRR285 pKa = 11.84DD286 pKa = 3.79VVEE289 pKa = 4.17YY290 pKa = 8.22TTDD293 pKa = 3.1ALNRR297 pKa = 11.84GLKK300 pKa = 9.91PLVLIDD306 pKa = 3.62IRR308 pKa = 11.84KK309 pKa = 9.96DD310 pKa = 3.09KK311 pKa = 10.7PKK313 pKa = 10.9NLNTPEE319 pKa = 4.16GAIEE323 pKa = 4.15WEE325 pKa = 4.36RR326 pKa = 11.84MVHH329 pKa = 7.07DD330 pKa = 5.4DD331 pKa = 3.76NNLIIDD337 pKa = 4.22MVNALDD343 pKa = 4.89KK344 pKa = 10.97RR345 pKa = 11.84VTVCAKK351 pKa = 10.06LRR353 pKa = 11.84PAFMQVGSMRR363 pKa = 11.84KK364 pKa = 8.67LLRR367 pKa = 11.84PVRR370 pKa = 11.84ILPLPYY376 pKa = 9.98LRR378 pKa = 11.84RR379 pKa = 11.84STAEE383 pKa = 3.44FNMFVPNEE391 pKa = 3.6ALMNGNEE398 pKa = 4.45IYY400 pKa = 10.7DD401 pKa = 3.58VTYY404 pKa = 11.16DD405 pKa = 3.53DD406 pKa = 4.63LVRR409 pKa = 11.84MSSEE413 pKa = 3.93VFVLKK418 pKa = 10.86NIIGGLYY425 pKa = 10.64NMYY428 pKa = 10.47LKK430 pKa = 10.85DD431 pKa = 3.22MHH433 pKa = 7.11LNLGVVNKK441 pKa = 10.26SVSLSDD447 pKa = 3.37GSSAIWSLSNINNEE461 pKa = 4.25RR462 pKa = 11.84ISNFNFNNFLYY473 pKa = 10.33AAPYY477 pKa = 10.6SDD479 pKa = 4.27FATSSVKK486 pKa = 10.39RR487 pKa = 11.84HH488 pKa = 4.98FKK490 pKa = 9.63GRR492 pKa = 11.84NYY494 pKa = 10.74SDD496 pKa = 2.98WCLNILDD503 pKa = 4.57EE504 pKa = 4.76VNLKK508 pKa = 10.88DD509 pKa = 4.12GVYY512 pKa = 10.25LVPLYY517 pKa = 10.79AIVGGGQITSHH528 pKa = 6.86DD529 pKa = 4.03FVNAIITDD537 pKa = 3.74QEE539 pKa = 3.96QLIDD543 pKa = 3.71FTQSEE548 pKa = 4.86RR549 pKa = 11.84ALSTQVVKK557 pKa = 10.75LVSFILKK564 pKa = 10.48DD565 pKa = 3.34SFTAKK570 pKa = 9.98GLNWTEE576 pKa = 3.29IDD578 pKa = 3.35NEE580 pKa = 3.79IRR582 pKa = 11.84NRR584 pKa = 11.84RR585 pKa = 11.84LSSLSGVGFTVTKK598 pKa = 10.1MLDD601 pKa = 3.34GKK603 pKa = 10.59VLVDD607 pKa = 3.66GKK609 pKa = 10.72VVTVSGHH616 pKa = 4.53MLYY619 pKa = 10.1ILLGSILGLPYY630 pKa = 10.6GIKK633 pKa = 10.13KK634 pKa = 9.14YY635 pKa = 10.78LKK637 pKa = 9.99EE638 pKa = 3.99IEE640 pKa = 4.44LNILKK645 pKa = 10.07PGSSYY650 pKa = 10.95EE651 pKa = 3.93RR652 pKa = 11.84GVGGRR657 pKa = 11.84VWHH660 pKa = 6.14GLISHH665 pKa = 6.58YY666 pKa = 10.79LAVDD670 pKa = 3.59CVIDD674 pKa = 4.55VIDD677 pKa = 4.4EE678 pKa = 4.4YY679 pKa = 10.7MVCTYY684 pKa = 10.44EE685 pKa = 5.62DD686 pKa = 3.62RR687 pKa = 11.84SKK689 pKa = 11.58LNVVLRR695 pKa = 11.84YY696 pKa = 9.79VKK698 pKa = 10.64SKK700 pKa = 10.72LLEE703 pKa = 4.63LGSKK707 pKa = 9.79YY708 pKa = 10.61DD709 pKa = 3.28VYY711 pKa = 11.43LSVDD715 pKa = 3.18EE716 pKa = 4.78RR717 pKa = 11.84LVLL720 pKa = 4.14
Molecular weight: 82.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5924 |
180 |
1214 |
493.7 |
55.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.313 ± 0.493 | 1.266 ± 0.201 |
6.6 ± 0.311 | 5.334 ± 0.208 |
3.764 ± 0.281 | 5.385 ± 0.302 |
2.296 ± 0.211 | 6.769 ± 0.266 |
6.617 ± 0.347 | 9.217 ± 0.332 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.684 ± 0.102 | 6.769 ± 0.267 |
3.309 ± 0.373 | 3.022 ± 0.388 |
4.693 ± 0.333 | 7.36 ± 0.363 |
5.807 ± 0.418 | 7.917 ± 0.268 |
0.608 ± 0.121 | 4.271 ± 0.223 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
