
Measles virus (strain Edmonston) (MeV) (Subacute sclerose panencephalitis virus)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Orthoparamyxovirinae; Morbillivirus; Measles morbillivirus
Average proteome isoelectric point is 7.02
Get precalculated fractions of proteins
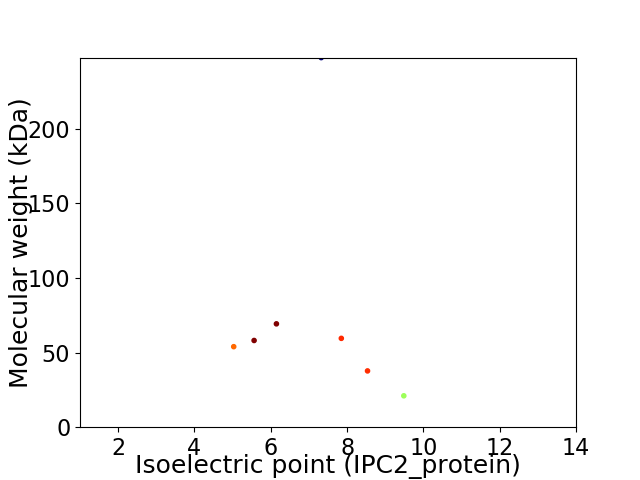
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|Q76VZ7|Q76VZ7_MEASE Phosphoprotein OS=Measles virus (strain Edmonston) OX=11235 PE=3 SV=1
MM1 pKa = 8.15AEE3 pKa = 3.63EE4 pKa = 3.91QARR7 pKa = 11.84HH8 pKa = 4.84VKK10 pKa = 10.61NGLEE14 pKa = 4.48CIRR17 pKa = 11.84ALKK20 pKa = 10.54AEE22 pKa = 5.1PIGSLAIEE30 pKa = 4.18EE31 pKa = 5.21AMAAWSEE38 pKa = 4.06ISDD41 pKa = 3.59NPGQEE46 pKa = 3.43RR47 pKa = 11.84ATCRR51 pKa = 11.84EE52 pKa = 4.01EE53 pKa = 4.01KK54 pKa = 10.5AGSSGLSKK62 pKa = 10.51PCLSAIGSTEE72 pKa = 3.52GGAPRR77 pKa = 11.84IRR79 pKa = 11.84GQGPGEE85 pKa = 4.18SDD87 pKa = 4.51DD88 pKa = 4.25DD89 pKa = 4.84AEE91 pKa = 4.54TLGIPPRR98 pKa = 11.84NLQASSTGLQCYY110 pKa = 8.82YY111 pKa = 10.96VYY113 pKa = 10.8DD114 pKa = 3.84HH115 pKa = 6.99SGEE118 pKa = 4.12AVKK121 pKa = 10.71GIQDD125 pKa = 3.6ADD127 pKa = 3.98SIMVQSGLDD136 pKa = 3.52GDD138 pKa = 4.4STLSGGDD145 pKa = 3.54NEE147 pKa = 5.15SEE149 pKa = 4.21NSDD152 pKa = 3.37VDD154 pKa = 3.07IGEE157 pKa = 4.99PDD159 pKa = 3.41TEE161 pKa = 4.81GYY163 pKa = 10.86AITDD167 pKa = 3.66RR168 pKa = 11.84GSAPISMGFRR178 pKa = 11.84ASDD181 pKa = 3.44VEE183 pKa = 4.22TAEE186 pKa = 4.19GGEE189 pKa = 3.83IHH191 pKa = 6.91EE192 pKa = 4.65LLRR195 pKa = 11.84LQSRR199 pKa = 11.84GNNFPKK205 pKa = 10.39LGKK208 pKa = 7.92TLNVPPPPDD217 pKa = 4.04PGRR220 pKa = 11.84ASTSGTPIKK229 pKa = 10.51KK230 pKa = 8.78GTDD233 pKa = 2.83ARR235 pKa = 11.84LASFGTEE242 pKa = 3.4IASLLTGGATQCARR256 pKa = 11.84KK257 pKa = 9.94SPSEE261 pKa = 3.79PSGPGAPAGNVPEE274 pKa = 4.59CVSNAALIQEE284 pKa = 4.51WTPEE288 pKa = 4.01SGTTISPRR296 pKa = 11.84SQNNEE301 pKa = 3.77EE302 pKa = 4.15GGDD305 pKa = 3.84YY306 pKa = 11.05YY307 pKa = 11.31DD308 pKa = 5.98DD309 pKa = 4.14EE310 pKa = 5.28LFSDD314 pKa = 3.92VQDD317 pKa = 3.6IKK319 pKa = 10.35TALAKK324 pKa = 10.19IHH326 pKa = 7.01EE327 pKa = 5.15DD328 pKa = 3.2NQKK331 pKa = 10.34IISKK335 pKa = 10.59LEE337 pKa = 3.76SLLLLKK343 pKa = 11.16GEE345 pKa = 4.15VEE347 pKa = 4.55SIKK350 pKa = 10.81KK351 pKa = 9.72QINRR355 pKa = 11.84QNISISTLEE364 pKa = 3.97GHH366 pKa = 7.29LSSIMIAIPGLGKK379 pKa = 10.47DD380 pKa = 4.25PNDD383 pKa = 3.35PTADD387 pKa = 3.52VEE389 pKa = 4.53INPDD393 pKa = 3.86LKK395 pKa = 10.89PIIGRR400 pKa = 11.84DD401 pKa = 3.18SGRR404 pKa = 11.84ALAEE408 pKa = 4.04VLKK411 pKa = 10.88KK412 pKa = 10.12PVASRR417 pKa = 11.84QLQGMTNGRR426 pKa = 11.84TSSRR430 pKa = 11.84GQLLKK435 pKa = 10.65EE436 pKa = 4.19FQLKK440 pKa = 10.35PIGKK444 pKa = 9.26KK445 pKa = 9.07MSSAVGFVPDD455 pKa = 4.19TGPASRR461 pKa = 11.84SVIRR465 pKa = 11.84SIIKK469 pKa = 10.0SSRR472 pKa = 11.84LEE474 pKa = 3.95EE475 pKa = 3.99DD476 pKa = 2.97RR477 pKa = 11.84KK478 pKa = 10.54RR479 pKa = 11.84YY480 pKa = 10.31LMTLLDD486 pKa = 5.15DD487 pKa = 4.11IKK489 pKa = 11.13GANDD493 pKa = 3.38LAKK496 pKa = 10.19FHH498 pKa = 6.04QMLMKK503 pKa = 10.09IIMKK507 pKa = 10.21
MM1 pKa = 8.15AEE3 pKa = 3.63EE4 pKa = 3.91QARR7 pKa = 11.84HH8 pKa = 4.84VKK10 pKa = 10.61NGLEE14 pKa = 4.48CIRR17 pKa = 11.84ALKK20 pKa = 10.54AEE22 pKa = 5.1PIGSLAIEE30 pKa = 4.18EE31 pKa = 5.21AMAAWSEE38 pKa = 4.06ISDD41 pKa = 3.59NPGQEE46 pKa = 3.43RR47 pKa = 11.84ATCRR51 pKa = 11.84EE52 pKa = 4.01EE53 pKa = 4.01KK54 pKa = 10.5AGSSGLSKK62 pKa = 10.51PCLSAIGSTEE72 pKa = 3.52GGAPRR77 pKa = 11.84IRR79 pKa = 11.84GQGPGEE85 pKa = 4.18SDD87 pKa = 4.51DD88 pKa = 4.25DD89 pKa = 4.84AEE91 pKa = 4.54TLGIPPRR98 pKa = 11.84NLQASSTGLQCYY110 pKa = 8.82YY111 pKa = 10.96VYY113 pKa = 10.8DD114 pKa = 3.84HH115 pKa = 6.99SGEE118 pKa = 4.12AVKK121 pKa = 10.71GIQDD125 pKa = 3.6ADD127 pKa = 3.98SIMVQSGLDD136 pKa = 3.52GDD138 pKa = 4.4STLSGGDD145 pKa = 3.54NEE147 pKa = 5.15SEE149 pKa = 4.21NSDD152 pKa = 3.37VDD154 pKa = 3.07IGEE157 pKa = 4.99PDD159 pKa = 3.41TEE161 pKa = 4.81GYY163 pKa = 10.86AITDD167 pKa = 3.66RR168 pKa = 11.84GSAPISMGFRR178 pKa = 11.84ASDD181 pKa = 3.44VEE183 pKa = 4.22TAEE186 pKa = 4.19GGEE189 pKa = 3.83IHH191 pKa = 6.91EE192 pKa = 4.65LLRR195 pKa = 11.84LQSRR199 pKa = 11.84GNNFPKK205 pKa = 10.39LGKK208 pKa = 7.92TLNVPPPPDD217 pKa = 4.04PGRR220 pKa = 11.84ASTSGTPIKK229 pKa = 10.51KK230 pKa = 8.78GTDD233 pKa = 2.83ARR235 pKa = 11.84LASFGTEE242 pKa = 3.4IASLLTGGATQCARR256 pKa = 11.84KK257 pKa = 9.94SPSEE261 pKa = 3.79PSGPGAPAGNVPEE274 pKa = 4.59CVSNAALIQEE284 pKa = 4.51WTPEE288 pKa = 4.01SGTTISPRR296 pKa = 11.84SQNNEE301 pKa = 3.77EE302 pKa = 4.15GGDD305 pKa = 3.84YY306 pKa = 11.05YY307 pKa = 11.31DD308 pKa = 5.98DD309 pKa = 4.14EE310 pKa = 5.28LFSDD314 pKa = 3.92VQDD317 pKa = 3.6IKK319 pKa = 10.35TALAKK324 pKa = 10.19IHH326 pKa = 7.01EE327 pKa = 5.15DD328 pKa = 3.2NQKK331 pKa = 10.34IISKK335 pKa = 10.59LEE337 pKa = 3.76SLLLLKK343 pKa = 11.16GEE345 pKa = 4.15VEE347 pKa = 4.55SIKK350 pKa = 10.81KK351 pKa = 9.72QINRR355 pKa = 11.84QNISISTLEE364 pKa = 3.97GHH366 pKa = 7.29LSSIMIAIPGLGKK379 pKa = 10.47DD380 pKa = 4.25PNDD383 pKa = 3.35PTADD387 pKa = 3.52VEE389 pKa = 4.53INPDD393 pKa = 3.86LKK395 pKa = 10.89PIIGRR400 pKa = 11.84DD401 pKa = 3.18SGRR404 pKa = 11.84ALAEE408 pKa = 4.04VLKK411 pKa = 10.88KK412 pKa = 10.12PVASRR417 pKa = 11.84QLQGMTNGRR426 pKa = 11.84TSSRR430 pKa = 11.84GQLLKK435 pKa = 10.65EE436 pKa = 4.19FQLKK440 pKa = 10.35PIGKK444 pKa = 9.26KK445 pKa = 9.07MSSAVGFVPDD455 pKa = 4.19TGPASRR461 pKa = 11.84SVIRR465 pKa = 11.84SIIKK469 pKa = 10.0SSRR472 pKa = 11.84LEE474 pKa = 3.95EE475 pKa = 3.99DD476 pKa = 2.97RR477 pKa = 11.84KK478 pKa = 10.54RR479 pKa = 11.84YY480 pKa = 10.31LMTLLDD486 pKa = 5.15DD487 pKa = 4.11IKK489 pKa = 11.13GANDD493 pKa = 3.38LAKK496 pKa = 10.19FHH498 pKa = 6.04QMLMKK503 pKa = 10.09IIMKK507 pKa = 10.21
Molecular weight: 53.96 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q76VZ7|Q76VZ7_MEASE Phosphoprotein OS=Measles virus (strain Edmonston) OX=11235 PE=3 SV=1
MM1 pKa = 7.9SKK3 pKa = 9.54TDD5 pKa = 3.3WNASGLSRR13 pKa = 11.84PSPSAHH19 pKa = 5.35WPSRR23 pKa = 11.84KK24 pKa = 9.12LWQHH28 pKa = 4.13GQKK31 pKa = 10.36YY32 pKa = 7.22QTTQDD37 pKa = 3.24RR38 pKa = 11.84SEE40 pKa = 4.24PPAGKK45 pKa = 9.52RR46 pKa = 11.84RR47 pKa = 11.84QAVRR51 pKa = 11.84VSANHH56 pKa = 7.14ASQQLDD62 pKa = 3.29QLKK65 pKa = 10.12AVHH68 pKa = 6.38LASAVRR74 pKa = 11.84DD75 pKa = 3.66LEE77 pKa = 4.45RR78 pKa = 11.84AMTTLKK84 pKa = 10.51LWEE87 pKa = 4.53SPQEE91 pKa = 4.11ISRR94 pKa = 11.84HH95 pKa = 4.01QALGYY100 pKa = 9.1SVIMFMITAVKK111 pKa = 10.1RR112 pKa = 11.84LRR114 pKa = 11.84EE115 pKa = 4.28SKK117 pKa = 10.12MLTLSWFNQALMVIAPSQEE136 pKa = 3.32EE137 pKa = 4.5TMNLKK142 pKa = 8.49TAMWILANLIPRR154 pKa = 11.84DD155 pKa = 4.0MLSLTGDD162 pKa = 4.77LLPSLWGSGLLMLKK176 pKa = 9.76LQKK179 pKa = 10.13EE180 pKa = 4.71GRR182 pKa = 11.84STSSS186 pKa = 2.66
MM1 pKa = 7.9SKK3 pKa = 9.54TDD5 pKa = 3.3WNASGLSRR13 pKa = 11.84PSPSAHH19 pKa = 5.35WPSRR23 pKa = 11.84KK24 pKa = 9.12LWQHH28 pKa = 4.13GQKK31 pKa = 10.36YY32 pKa = 7.22QTTQDD37 pKa = 3.24RR38 pKa = 11.84SEE40 pKa = 4.24PPAGKK45 pKa = 9.52RR46 pKa = 11.84RR47 pKa = 11.84QAVRR51 pKa = 11.84VSANHH56 pKa = 7.14ASQQLDD62 pKa = 3.29QLKK65 pKa = 10.12AVHH68 pKa = 6.38LASAVRR74 pKa = 11.84DD75 pKa = 3.66LEE77 pKa = 4.45RR78 pKa = 11.84AMTTLKK84 pKa = 10.51LWEE87 pKa = 4.53SPQEE91 pKa = 4.11ISRR94 pKa = 11.84HH95 pKa = 4.01QALGYY100 pKa = 9.1SVIMFMITAVKK111 pKa = 10.1RR112 pKa = 11.84LRR114 pKa = 11.84EE115 pKa = 4.28SKK117 pKa = 10.12MLTLSWFNQALMVIAPSQEE136 pKa = 3.32EE137 pKa = 4.5TMNLKK142 pKa = 8.49TAMWILANLIPRR154 pKa = 11.84DD155 pKa = 4.0MLSLTGDD162 pKa = 4.77LLPSLWGSGLLMLKK176 pKa = 9.76LQKK179 pKa = 10.13EE180 pKa = 4.71GRR182 pKa = 11.84STSSS186 pKa = 2.66
Molecular weight: 21.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4901 |
186 |
2183 |
700.1 |
78.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.08 ± 0.535 | 1.694 ± 0.262 |
5.366 ± 0.408 | 5.468 ± 0.475 |
3.244 ± 0.511 | 6.754 ± 0.761 |
2.265 ± 0.523 | 7.284 ± 0.434 |
5.142 ± 0.301 | 10.855 ± 0.477 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.571 ± 0.176 | 4.162 ± 0.249 |
4.754 ± 0.431 | 3.673 ± 0.369 |
5.958 ± 0.328 | 8.508 ± 0.444 |
5.55 ± 0.149 | 6.182 ± 0.57 |
1.102 ± 0.206 | 3.387 ± 0.501 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
