
Cellulomonas cellasea DSM 20118
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Cellulomonadaceae; Cellulomonas; Cellulomonas cellasea
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
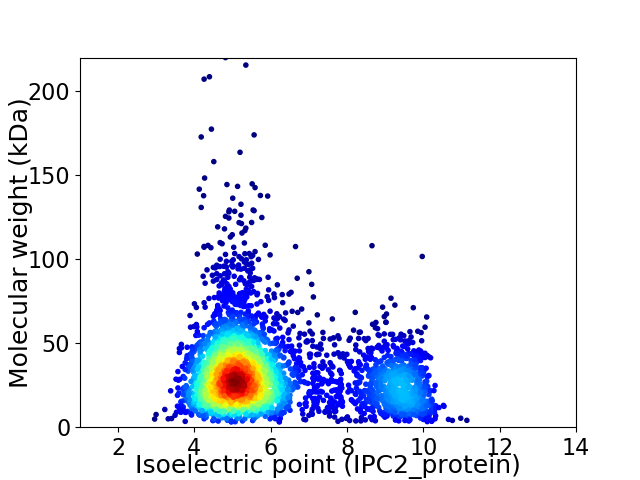
Virtual 2D-PAGE plot for 3560 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A0BBD4|A0A0A0BBD4_9CELL RNA polymerase subunit sigma-24 OS=Cellulomonas cellasea DSM 20118 OX=1408250 GN=Q760_12500 PE=3 SV=1
MM1 pKa = 7.21NGMRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84GTAVVAAGAGLTLLLAACSGGGDD31 pKa = 3.52EE32 pKa = 5.58GGGDD36 pKa = 3.87AEE38 pKa = 4.55GTANEE43 pKa = 4.49TDD45 pKa = 3.08CSAYY49 pKa = 10.11EE50 pKa = 4.07EE51 pKa = 4.46YY52 pKa = 11.32GDD54 pKa = 4.89LSGKK58 pKa = 7.18TVSVYY63 pKa = 10.32TSIVDD68 pKa = 3.71PEE70 pKa = 4.27SEE72 pKa = 4.15EE73 pKa = 4.44QINSYY78 pKa = 10.53QPFVDD83 pKa = 3.94CTGVEE88 pKa = 3.61IDD90 pKa = 3.94YY91 pKa = 10.77EE92 pKa = 4.55GSRR95 pKa = 11.84EE96 pKa = 3.99FEE98 pKa = 3.84AQLPVRR104 pKa = 11.84LTAGNPPDD112 pKa = 3.5IAYY115 pKa = 9.99VPQPGFLKK123 pKa = 10.84SLVADD128 pKa = 5.0FPDD131 pKa = 3.46QVVPAPQSVIDD142 pKa = 4.12NATEE146 pKa = 4.86FYY148 pKa = 10.33TEE150 pKa = 3.49EE151 pKa = 3.86WVNYY155 pKa = 7.14GTVDD159 pKa = 3.13GTLYY163 pKa = 9.35ATPLGANVKK172 pKa = 10.15SFVWYY177 pKa = 10.41SPLAFEE183 pKa = 4.87DD184 pKa = 3.29AGYY187 pKa = 9.64EE188 pKa = 4.69IPTTWDD194 pKa = 2.95EE195 pKa = 5.71LMDD198 pKa = 4.45LSQQIVDD205 pKa = 4.07DD206 pKa = 5.79GGVPWCAGIEE216 pKa = 4.19SGDD219 pKa = 3.58ATGWPATDD227 pKa = 3.11WLEE230 pKa = 4.41DD231 pKa = 3.59VVLRR235 pKa = 11.84TAGADD240 pKa = 4.48VYY242 pKa = 10.24DD243 pKa = 3.09QWVNHH248 pKa = 6.96EE249 pKa = 4.64IPFNDD254 pKa = 3.74PQIVEE259 pKa = 4.14ALGTVGEE266 pKa = 4.09ILKK269 pKa = 10.38NDD271 pKa = 3.57EE272 pKa = 4.36YY273 pKa = 11.96VNGGLGNVQSIATAAWNEE291 pKa = 4.22SANGIPDD298 pKa = 4.05GTCWLHH304 pKa = 6.31RR305 pKa = 11.84AANFYY310 pKa = 8.96QANWDD315 pKa = 3.54EE316 pKa = 4.33SLEE319 pKa = 4.12VAEE322 pKa = 5.63DD323 pKa = 3.81GDD325 pKa = 4.2VYY327 pKa = 11.44AFYY330 pKa = 11.05LPGVSEE336 pKa = 4.61DD337 pKa = 4.24DD338 pKa = 4.18KK339 pKa = 11.21PLLGAGEE346 pKa = 4.4FVTAFSDD353 pKa = 3.8RR354 pKa = 11.84PEE356 pKa = 4.07VEE358 pKa = 3.94AFQAFLSSPEE368 pKa = 3.89WANSKK373 pKa = 10.93ASVTAQGWFSANNGLDD389 pKa = 3.4PEE391 pKa = 5.26LIKK394 pKa = 11.18SPIDD398 pKa = 3.26KK399 pKa = 10.59LAFEE403 pKa = 5.43LLTSDD408 pKa = 4.22EE409 pKa = 4.3YY410 pKa = 11.16TFRR413 pKa = 11.84FDD415 pKa = 6.12GSDD418 pKa = 3.49QMPGAVGTGSFWTEE432 pKa = 3.25MTAWVANDD440 pKa = 3.71KK441 pKa = 11.03SDD443 pKa = 3.9EE444 pKa = 4.26DD445 pKa = 3.72VLSAIEE451 pKa = 4.2ASWPTSS457 pKa = 3.11
MM1 pKa = 7.21NGMRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84GTAVVAAGAGLTLLLAACSGGGDD31 pKa = 3.52EE32 pKa = 5.58GGGDD36 pKa = 3.87AEE38 pKa = 4.55GTANEE43 pKa = 4.49TDD45 pKa = 3.08CSAYY49 pKa = 10.11EE50 pKa = 4.07EE51 pKa = 4.46YY52 pKa = 11.32GDD54 pKa = 4.89LSGKK58 pKa = 7.18TVSVYY63 pKa = 10.32TSIVDD68 pKa = 3.71PEE70 pKa = 4.27SEE72 pKa = 4.15EE73 pKa = 4.44QINSYY78 pKa = 10.53QPFVDD83 pKa = 3.94CTGVEE88 pKa = 3.61IDD90 pKa = 3.94YY91 pKa = 10.77EE92 pKa = 4.55GSRR95 pKa = 11.84EE96 pKa = 3.99FEE98 pKa = 3.84AQLPVRR104 pKa = 11.84LTAGNPPDD112 pKa = 3.5IAYY115 pKa = 9.99VPQPGFLKK123 pKa = 10.84SLVADD128 pKa = 5.0FPDD131 pKa = 3.46QVVPAPQSVIDD142 pKa = 4.12NATEE146 pKa = 4.86FYY148 pKa = 10.33TEE150 pKa = 3.49EE151 pKa = 3.86WVNYY155 pKa = 7.14GTVDD159 pKa = 3.13GTLYY163 pKa = 9.35ATPLGANVKK172 pKa = 10.15SFVWYY177 pKa = 10.41SPLAFEE183 pKa = 4.87DD184 pKa = 3.29AGYY187 pKa = 9.64EE188 pKa = 4.69IPTTWDD194 pKa = 2.95EE195 pKa = 5.71LMDD198 pKa = 4.45LSQQIVDD205 pKa = 4.07DD206 pKa = 5.79GGVPWCAGIEE216 pKa = 4.19SGDD219 pKa = 3.58ATGWPATDD227 pKa = 3.11WLEE230 pKa = 4.41DD231 pKa = 3.59VVLRR235 pKa = 11.84TAGADD240 pKa = 4.48VYY242 pKa = 10.24DD243 pKa = 3.09QWVNHH248 pKa = 6.96EE249 pKa = 4.64IPFNDD254 pKa = 3.74PQIVEE259 pKa = 4.14ALGTVGEE266 pKa = 4.09ILKK269 pKa = 10.38NDD271 pKa = 3.57EE272 pKa = 4.36YY273 pKa = 11.96VNGGLGNVQSIATAAWNEE291 pKa = 4.22SANGIPDD298 pKa = 4.05GTCWLHH304 pKa = 6.31RR305 pKa = 11.84AANFYY310 pKa = 8.96QANWDD315 pKa = 3.54EE316 pKa = 4.33SLEE319 pKa = 4.12VAEE322 pKa = 5.63DD323 pKa = 3.81GDD325 pKa = 4.2VYY327 pKa = 11.44AFYY330 pKa = 11.05LPGVSEE336 pKa = 4.61DD337 pKa = 4.24DD338 pKa = 4.18KK339 pKa = 11.21PLLGAGEE346 pKa = 4.4FVTAFSDD353 pKa = 3.8RR354 pKa = 11.84PEE356 pKa = 4.07VEE358 pKa = 3.94AFQAFLSSPEE368 pKa = 3.89WANSKK373 pKa = 10.93ASVTAQGWFSANNGLDD389 pKa = 3.4PEE391 pKa = 5.26LIKK394 pKa = 11.18SPIDD398 pKa = 3.26KK399 pKa = 10.59LAFEE403 pKa = 5.43LLTSDD408 pKa = 4.22EE409 pKa = 4.3YY410 pKa = 11.16TFRR413 pKa = 11.84FDD415 pKa = 6.12GSDD418 pKa = 3.49QMPGAVGTGSFWTEE432 pKa = 3.25MTAWVANDD440 pKa = 3.71KK441 pKa = 11.03SDD443 pKa = 3.9EE444 pKa = 4.26DD445 pKa = 3.72VLSAIEE451 pKa = 4.2ASWPTSS457 pKa = 3.11
Molecular weight: 49.32 kDa
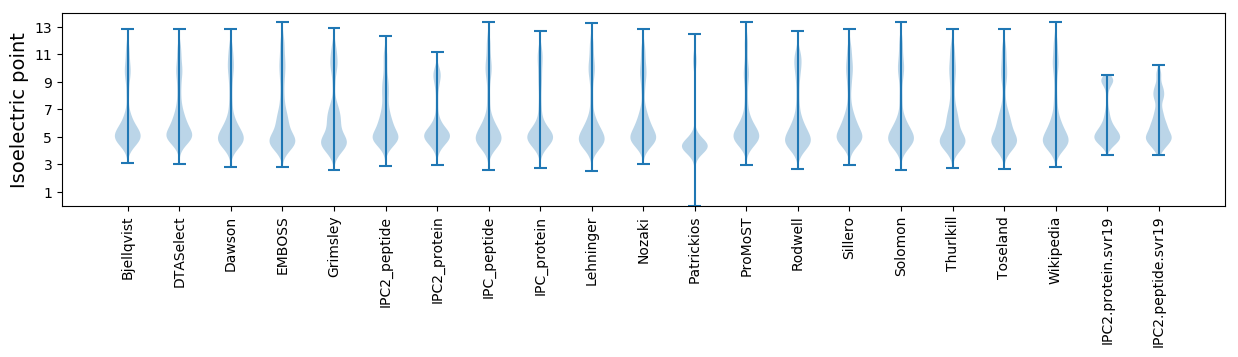
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A0B6P3|A0A0A0B6P3_9CELL Membrane protein OS=Cellulomonas cellasea DSM 20118 OX=1408250 GN=Q760_12970 PE=3 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1137429 |
29 |
2089 |
319.5 |
33.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.435 ± 0.061 | 0.57 ± 0.009 |
6.403 ± 0.036 | 5.371 ± 0.039 |
2.492 ± 0.022 | 9.593 ± 0.039 |
2.161 ± 0.023 | 2.682 ± 0.029 |
1.3 ± 0.026 | 10.369 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.464 ± 0.013 | 1.505 ± 0.025 |
6.042 ± 0.033 | 2.558 ± 0.022 |
7.917 ± 0.046 | 4.949 ± 0.029 |
6.576 ± 0.041 | 10.193 ± 0.049 |
1.577 ± 0.019 | 1.844 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
