
Baminivirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; unclassified Geminiviridae
Average proteome isoelectric point is 8.34
Get precalculated fractions of proteins
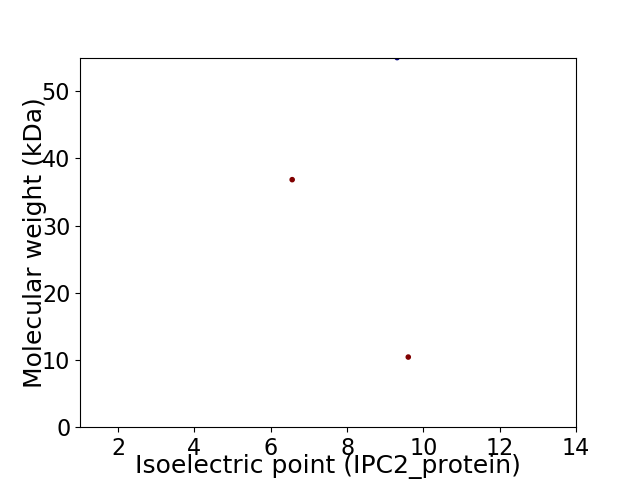
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
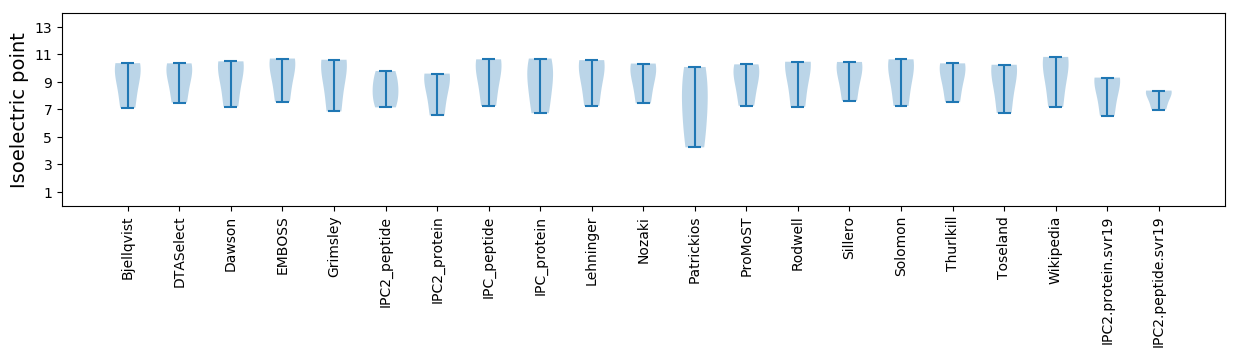
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|J9TCS3|J9TCS3_9GEMI Putative movement protein OS=Baminivirus OX=1229324 PE=4 SV=1
MM1 pKa = 7.1AQPRR5 pKa = 11.84FRR7 pKa = 11.84LQGKK11 pKa = 9.6NILLTYY17 pKa = 6.94PQCNLEE23 pKa = 4.26VKK25 pKa = 10.03HH26 pKa = 6.17VCDD29 pKa = 4.01EE30 pKa = 4.19LALRR34 pKa = 11.84LPPGRR39 pKa = 11.84CSGVLEE45 pKa = 4.09RR46 pKa = 11.84HH47 pKa = 5.56QDD49 pKa = 3.74GNPHH53 pKa = 6.88LHH55 pKa = 6.51ILWEE59 pKa = 3.94LRR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84LDD65 pKa = 3.34TQNPRR70 pKa = 11.84FFDD73 pKa = 3.03VDD75 pKa = 3.67TFHH78 pKa = 7.26PNVVVVRR85 pKa = 11.84SVRR88 pKa = 11.84GARR91 pKa = 11.84DD92 pKa = 3.53YY93 pKa = 10.76IAKK96 pKa = 9.2TGAEE100 pKa = 4.45VYY102 pKa = 9.72TNEE105 pKa = 4.42PGEE108 pKa = 4.36PEE110 pKa = 4.16CEE112 pKa = 3.86SWGEE116 pKa = 3.86ILEE119 pKa = 4.35QATSRR124 pKa = 11.84EE125 pKa = 3.98GFLDD129 pKa = 3.81LVRR132 pKa = 11.84KK133 pKa = 9.53HH134 pKa = 6.14KK135 pKa = 10.57PRR137 pKa = 11.84DD138 pKa = 3.62FILNMEE144 pKa = 4.36RR145 pKa = 11.84LEE147 pKa = 4.1YY148 pKa = 10.33AANILFAEE156 pKa = 4.82PEE158 pKa = 3.91QSFVSMYY165 pKa = 8.02TTFAPPSACHH175 pKa = 6.63DD176 pKa = 2.68WWLQYY181 pKa = 9.97GARR184 pKa = 11.84PRR186 pKa = 11.84LEE188 pKa = 4.34GRR190 pKa = 11.84LQSLYY195 pKa = 11.06VEE197 pKa = 4.66GASRR201 pKa = 11.84IGKK204 pKa = 6.82TEE206 pKa = 3.79WARR209 pKa = 11.84SLGPHH214 pKa = 6.12VYY216 pKa = 10.12FNNMYY221 pKa = 10.78NLDD224 pKa = 4.08DD225 pKa = 4.66LKK227 pKa = 11.3SALRR231 pKa = 11.84YY232 pKa = 10.36AEE234 pKa = 3.99FAVFDD239 pKa = 5.57DD240 pKa = 4.26IPFDD244 pKa = 5.0RR245 pKa = 11.84LPAWKK250 pKa = 10.39SFFGCQHH257 pKa = 6.68TFTLTDD263 pKa = 3.87RR264 pKa = 11.84YY265 pKa = 9.97RR266 pKa = 11.84KK267 pKa = 10.13KK268 pKa = 10.13FTISDD273 pKa = 3.41WSKK276 pKa = 10.72PSIFLVNPDD285 pKa = 3.31MTYY288 pKa = 10.96DD289 pKa = 2.99AWNKK293 pKa = 8.31GRR295 pKa = 11.84EE296 pKa = 3.97WEE298 pKa = 4.3YY299 pKa = 10.47IKK301 pKa = 11.02QNAVIVKK308 pKa = 8.48TDD310 pKa = 2.82SRR312 pKa = 11.84FYY314 pKa = 11.47
MM1 pKa = 7.1AQPRR5 pKa = 11.84FRR7 pKa = 11.84LQGKK11 pKa = 9.6NILLTYY17 pKa = 6.94PQCNLEE23 pKa = 4.26VKK25 pKa = 10.03HH26 pKa = 6.17VCDD29 pKa = 4.01EE30 pKa = 4.19LALRR34 pKa = 11.84LPPGRR39 pKa = 11.84CSGVLEE45 pKa = 4.09RR46 pKa = 11.84HH47 pKa = 5.56QDD49 pKa = 3.74GNPHH53 pKa = 6.88LHH55 pKa = 6.51ILWEE59 pKa = 3.94LRR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84LDD65 pKa = 3.34TQNPRR70 pKa = 11.84FFDD73 pKa = 3.03VDD75 pKa = 3.67TFHH78 pKa = 7.26PNVVVVRR85 pKa = 11.84SVRR88 pKa = 11.84GARR91 pKa = 11.84DD92 pKa = 3.53YY93 pKa = 10.76IAKK96 pKa = 9.2TGAEE100 pKa = 4.45VYY102 pKa = 9.72TNEE105 pKa = 4.42PGEE108 pKa = 4.36PEE110 pKa = 4.16CEE112 pKa = 3.86SWGEE116 pKa = 3.86ILEE119 pKa = 4.35QATSRR124 pKa = 11.84EE125 pKa = 3.98GFLDD129 pKa = 3.81LVRR132 pKa = 11.84KK133 pKa = 9.53HH134 pKa = 6.14KK135 pKa = 10.57PRR137 pKa = 11.84DD138 pKa = 3.62FILNMEE144 pKa = 4.36RR145 pKa = 11.84LEE147 pKa = 4.1YY148 pKa = 10.33AANILFAEE156 pKa = 4.82PEE158 pKa = 3.91QSFVSMYY165 pKa = 8.02TTFAPPSACHH175 pKa = 6.63DD176 pKa = 2.68WWLQYY181 pKa = 9.97GARR184 pKa = 11.84PRR186 pKa = 11.84LEE188 pKa = 4.34GRR190 pKa = 11.84LQSLYY195 pKa = 11.06VEE197 pKa = 4.66GASRR201 pKa = 11.84IGKK204 pKa = 6.82TEE206 pKa = 3.79WARR209 pKa = 11.84SLGPHH214 pKa = 6.12VYY216 pKa = 10.12FNNMYY221 pKa = 10.78NLDD224 pKa = 4.08DD225 pKa = 4.66LKK227 pKa = 11.3SALRR231 pKa = 11.84YY232 pKa = 10.36AEE234 pKa = 3.99FAVFDD239 pKa = 5.57DD240 pKa = 4.26IPFDD244 pKa = 5.0RR245 pKa = 11.84LPAWKK250 pKa = 10.39SFFGCQHH257 pKa = 6.68TFTLTDD263 pKa = 3.87RR264 pKa = 11.84YY265 pKa = 9.97RR266 pKa = 11.84KK267 pKa = 10.13KK268 pKa = 10.13FTISDD273 pKa = 3.41WSKK276 pKa = 10.72PSIFLVNPDD285 pKa = 3.31MTYY288 pKa = 10.96DD289 pKa = 2.99AWNKK293 pKa = 8.31GRR295 pKa = 11.84EE296 pKa = 3.97WEE298 pKa = 4.3YY299 pKa = 10.47IKK301 pKa = 11.02QNAVIVKK308 pKa = 8.48TDD310 pKa = 2.82SRR312 pKa = 11.84FYY314 pKa = 11.47
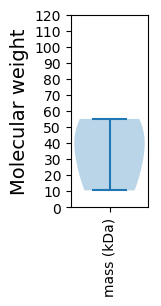
Molecular weight: 36.84 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|J9TCS3|J9TCS3_9GEMI Putative movement protein OS=Baminivirus OX=1229324 PE=4 SV=1
MM1 pKa = 7.98RR2 pKa = 11.84DD3 pKa = 3.21LRR5 pKa = 11.84DD6 pKa = 3.87FYY8 pKa = 11.59VPRR11 pKa = 11.84LRR13 pKa = 11.84MNEE16 pKa = 4.52FIRR19 pKa = 11.84RR20 pKa = 11.84PIGDD24 pKa = 4.08GLKK27 pKa = 10.19EE28 pKa = 4.21SPDD31 pKa = 3.6LCVLPGYY38 pKa = 10.26LGRR41 pKa = 11.84TNQVRR46 pKa = 11.84LSYY49 pKa = 10.08PGPTFTRR56 pKa = 11.84SVTRR60 pKa = 11.84SCPARR65 pKa = 11.84SRR67 pKa = 11.84DD68 pKa = 3.35PHH70 pKa = 7.97VIFLGWFGGAGGFTPQHH87 pKa = 6.19FLSIAA92 pKa = 3.25
MM1 pKa = 7.98RR2 pKa = 11.84DD3 pKa = 3.21LRR5 pKa = 11.84DD6 pKa = 3.87FYY8 pKa = 11.59VPRR11 pKa = 11.84LRR13 pKa = 11.84MNEE16 pKa = 4.52FIRR19 pKa = 11.84RR20 pKa = 11.84PIGDD24 pKa = 4.08GLKK27 pKa = 10.19EE28 pKa = 4.21SPDD31 pKa = 3.6LCVLPGYY38 pKa = 10.26LGRR41 pKa = 11.84TNQVRR46 pKa = 11.84LSYY49 pKa = 10.08PGPTFTRR56 pKa = 11.84SVTRR60 pKa = 11.84SCPARR65 pKa = 11.84SRR67 pKa = 11.84DD68 pKa = 3.35PHH70 pKa = 7.97VIFLGWFGGAGGFTPQHH87 pKa = 6.19FLSIAA92 pKa = 3.25
Molecular weight: 10.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
897 |
92 |
491 |
299.0 |
34.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.581 ± 1.249 | 1.226 ± 0.453 |
5.797 ± 0.116 | 4.125 ± 1.312 |
4.794 ± 1.002 | 7.246 ± 1.014 |
3.456 ± 0.464 | 4.125 ± 0.138 |
5.463 ± 1.182 | 8.361 ± 0.61 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.23 ± 0.32 | 3.79 ± 0.357 |
4.905 ± 1.327 | 3.567 ± 0.297 |
8.027 ± 1.164 | 6.466 ± 0.672 |
6.912 ± 1.209 | 5.351 ± 0.196 |
2.118 ± 0.342 | 4.459 ± 0.244 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
