
Aminobacter sp. DSM 101952
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Phyllobacteriaceae; Aminobacter; unclassified Aminobacter
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
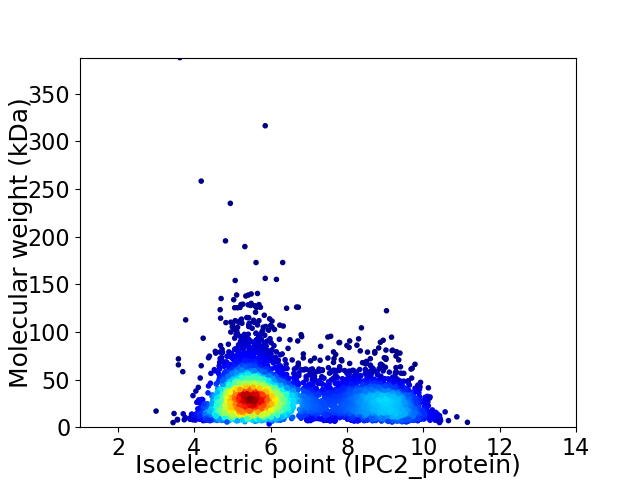
Virtual 2D-PAGE plot for 4851 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q6LLH8|A0A0Q6LLH8_9RHIZ Spermidine/putrescine ABC transporter permease OS=Aminobacter sp. DSM 101952 OX=2735891 GN=ASC75_12690 PE=3 SV=1
MM1 pKa = 6.6YY2 pKa = 10.05AAYY5 pKa = 9.92FDD7 pKa = 5.29RR8 pKa = 11.84ISEE11 pKa = 4.52TIAHH15 pKa = 7.09FIGLFDD21 pKa = 3.82TASEE25 pKa = 4.1EE26 pKa = 4.19ARR28 pKa = 11.84LKK30 pKa = 9.74QAYY33 pKa = 10.06DD34 pKa = 3.3EE35 pKa = 4.95FRR37 pKa = 11.84AKK39 pKa = 9.91QQADD43 pKa = 3.29EE44 pKa = 4.95HH45 pKa = 7.83LPEE48 pKa = 4.87HH49 pKa = 5.36QTAALKK55 pKa = 10.7VDD57 pKa = 3.82TSHH60 pKa = 6.65TLEE63 pKa = 5.38DD64 pKa = 4.32FDD66 pKa = 5.87PSLKK70 pKa = 10.44YY71 pKa = 10.64VGVRR75 pKa = 11.84PEE77 pKa = 3.88IEE79 pKa = 4.42QITVWSKK86 pKa = 10.83VAFTPPDD93 pKa = 3.3VAVPDD98 pKa = 4.41GPFLQNYY105 pKa = 8.31AHH107 pKa = 7.43LPQGTVGSGGIVLPQILPPGDD128 pKa = 3.27LAVIVNQEE136 pKa = 3.56IRR138 pKa = 11.84LSDD141 pKa = 3.62DD142 pKa = 3.84DD143 pKa = 4.14YY144 pKa = 11.96VGFGGSGLKK153 pKa = 10.3FNAPPVDD160 pKa = 4.28DD161 pKa = 4.42SPLVEE166 pKa = 4.29MQQAAADD173 pKa = 4.46LSPIDD178 pKa = 5.51DD179 pKa = 3.94LTLPGSTEE187 pKa = 3.64EE188 pKa = 3.99MATFVTTAQARR199 pKa = 11.84FNDD202 pKa = 4.13FDD204 pKa = 6.66ADD206 pKa = 3.64DD207 pKa = 4.58HH208 pKa = 6.84GNANVFCVKK217 pKa = 10.69AEE219 pKa = 4.4TIEE222 pKa = 3.96GTYY225 pKa = 10.68FNGQLIDD232 pKa = 3.67EE233 pKa = 4.71ADD235 pKa = 3.56APKK238 pKa = 10.11MEE240 pKa = 5.64DD241 pKa = 3.31YY242 pKa = 11.29INFLKK247 pKa = 10.87EE248 pKa = 3.52QDD250 pKa = 4.51DD251 pKa = 4.07DD252 pKa = 5.04AKK254 pKa = 11.47GPDD257 pKa = 3.57APLDD261 pKa = 3.48VDD263 pKa = 4.33TPPDD267 pKa = 3.5SSDD270 pKa = 5.32FMDD273 pKa = 3.34GWGDD277 pKa = 3.4GTANPSVEE285 pKa = 3.74ISTGGNTVINNAVVVNDD302 pKa = 3.8WASAEE307 pKa = 3.98VMAVMGDD314 pKa = 4.12HH315 pKa = 6.91ISLNAIIQINGTCDD329 pKa = 3.51ADD331 pKa = 3.9SVGAAVGGWPFDD343 pKa = 4.06PGGGDD348 pKa = 3.25NNLSFNIAMFKK359 pKa = 10.7HH360 pKa = 6.58IDD362 pKa = 3.46TSGGDD367 pKa = 3.47QAPAEE372 pKa = 4.51APTGFPAYY380 pKa = 10.24YY381 pKa = 9.97VVTEE385 pKa = 4.08ISGDD389 pKa = 3.88LIMMNWIEE397 pKa = 4.12QYY399 pKa = 11.74AFVTDD404 pKa = 3.66NDD406 pKa = 3.73VVIASSSGVQSVVSTGDD423 pKa = 3.02NTAFNGLSLNEE434 pKa = 3.71IGNYY438 pKa = 9.02YY439 pKa = 10.61DD440 pKa = 5.58LIIIGGNVYY449 pKa = 10.18DD450 pKa = 4.84ASIIHH455 pKa = 6.28QINLLLDD462 pKa = 3.64NDD464 pKa = 4.18VIGAVSGFEE473 pKa = 4.16TTGEE477 pKa = 4.37GSVSTGDD484 pKa = 3.38NLLWNQASIVNVGNATAEE502 pKa = 4.1ALPDD506 pKa = 4.43GYY508 pKa = 11.52NKK510 pKa = 10.59AAGDD514 pKa = 3.64LAAGNKK520 pKa = 7.82EE521 pKa = 4.21LPDD524 pKa = 5.56SILQDD529 pKa = 3.17AAFAGIGVLRR539 pKa = 11.84VLYY542 pKa = 10.32ISGDD546 pKa = 3.49IYY548 pKa = 10.95NLNYY552 pKa = 9.86IKK554 pKa = 9.57QTTVVGDD561 pKa = 3.66SDD563 pKa = 4.13QVALAMNNLVANPDD577 pKa = 3.88AEE579 pKa = 4.4WTITTGGNQLVNDD592 pKa = 4.06ATIVDD597 pKa = 3.63VDD599 pKa = 4.17GTSKK603 pKa = 10.38IYY605 pKa = 10.93VGGDD609 pKa = 3.21SYY611 pKa = 11.88SDD613 pKa = 3.97EE614 pKa = 4.37ILIQANLVSSEE625 pKa = 4.32PDD627 pKa = 3.08LGAQNPDD634 pKa = 2.94ILVTEE639 pKa = 4.3AVVFLDD645 pKa = 5.53DD646 pKa = 5.33GGDD649 pKa = 3.76DD650 pKa = 3.94TGDD653 pKa = 3.38ASNTNPIAPQPADD666 pKa = 3.53SGSADD671 pKa = 3.45VMQHH675 pKa = 5.04MLGG678 pKa = 3.58
MM1 pKa = 6.6YY2 pKa = 10.05AAYY5 pKa = 9.92FDD7 pKa = 5.29RR8 pKa = 11.84ISEE11 pKa = 4.52TIAHH15 pKa = 7.09FIGLFDD21 pKa = 3.82TASEE25 pKa = 4.1EE26 pKa = 4.19ARR28 pKa = 11.84LKK30 pKa = 9.74QAYY33 pKa = 10.06DD34 pKa = 3.3EE35 pKa = 4.95FRR37 pKa = 11.84AKK39 pKa = 9.91QQADD43 pKa = 3.29EE44 pKa = 4.95HH45 pKa = 7.83LPEE48 pKa = 4.87HH49 pKa = 5.36QTAALKK55 pKa = 10.7VDD57 pKa = 3.82TSHH60 pKa = 6.65TLEE63 pKa = 5.38DD64 pKa = 4.32FDD66 pKa = 5.87PSLKK70 pKa = 10.44YY71 pKa = 10.64VGVRR75 pKa = 11.84PEE77 pKa = 3.88IEE79 pKa = 4.42QITVWSKK86 pKa = 10.83VAFTPPDD93 pKa = 3.3VAVPDD98 pKa = 4.41GPFLQNYY105 pKa = 8.31AHH107 pKa = 7.43LPQGTVGSGGIVLPQILPPGDD128 pKa = 3.27LAVIVNQEE136 pKa = 3.56IRR138 pKa = 11.84LSDD141 pKa = 3.62DD142 pKa = 3.84DD143 pKa = 4.14YY144 pKa = 11.96VGFGGSGLKK153 pKa = 10.3FNAPPVDD160 pKa = 4.28DD161 pKa = 4.42SPLVEE166 pKa = 4.29MQQAAADD173 pKa = 4.46LSPIDD178 pKa = 5.51DD179 pKa = 3.94LTLPGSTEE187 pKa = 3.64EE188 pKa = 3.99MATFVTTAQARR199 pKa = 11.84FNDD202 pKa = 4.13FDD204 pKa = 6.66ADD206 pKa = 3.64DD207 pKa = 4.58HH208 pKa = 6.84GNANVFCVKK217 pKa = 10.69AEE219 pKa = 4.4TIEE222 pKa = 3.96GTYY225 pKa = 10.68FNGQLIDD232 pKa = 3.67EE233 pKa = 4.71ADD235 pKa = 3.56APKK238 pKa = 10.11MEE240 pKa = 5.64DD241 pKa = 3.31YY242 pKa = 11.29INFLKK247 pKa = 10.87EE248 pKa = 3.52QDD250 pKa = 4.51DD251 pKa = 4.07DD252 pKa = 5.04AKK254 pKa = 11.47GPDD257 pKa = 3.57APLDD261 pKa = 3.48VDD263 pKa = 4.33TPPDD267 pKa = 3.5SSDD270 pKa = 5.32FMDD273 pKa = 3.34GWGDD277 pKa = 3.4GTANPSVEE285 pKa = 3.74ISTGGNTVINNAVVVNDD302 pKa = 3.8WASAEE307 pKa = 3.98VMAVMGDD314 pKa = 4.12HH315 pKa = 6.91ISLNAIIQINGTCDD329 pKa = 3.51ADD331 pKa = 3.9SVGAAVGGWPFDD343 pKa = 4.06PGGGDD348 pKa = 3.25NNLSFNIAMFKK359 pKa = 10.7HH360 pKa = 6.58IDD362 pKa = 3.46TSGGDD367 pKa = 3.47QAPAEE372 pKa = 4.51APTGFPAYY380 pKa = 10.24YY381 pKa = 9.97VVTEE385 pKa = 4.08ISGDD389 pKa = 3.88LIMMNWIEE397 pKa = 4.12QYY399 pKa = 11.74AFVTDD404 pKa = 3.66NDD406 pKa = 3.73VVIASSSGVQSVVSTGDD423 pKa = 3.02NTAFNGLSLNEE434 pKa = 3.71IGNYY438 pKa = 9.02YY439 pKa = 10.61DD440 pKa = 5.58LIIIGGNVYY449 pKa = 10.18DD450 pKa = 4.84ASIIHH455 pKa = 6.28QINLLLDD462 pKa = 3.64NDD464 pKa = 4.18VIGAVSGFEE473 pKa = 4.16TTGEE477 pKa = 4.37GSVSTGDD484 pKa = 3.38NLLWNQASIVNVGNATAEE502 pKa = 4.1ALPDD506 pKa = 4.43GYY508 pKa = 11.52NKK510 pKa = 10.59AAGDD514 pKa = 3.64LAAGNKK520 pKa = 7.82EE521 pKa = 4.21LPDD524 pKa = 5.56SILQDD529 pKa = 3.17AAFAGIGVLRR539 pKa = 11.84VLYY542 pKa = 10.32ISGDD546 pKa = 3.49IYY548 pKa = 10.95NLNYY552 pKa = 9.86IKK554 pKa = 9.57QTTVVGDD561 pKa = 3.66SDD563 pKa = 4.13QVALAMNNLVANPDD577 pKa = 3.88AEE579 pKa = 4.4WTITTGGNQLVNDD592 pKa = 4.06ATIVDD597 pKa = 3.63VDD599 pKa = 4.17GTSKK603 pKa = 10.38IYY605 pKa = 10.93VGGDD609 pKa = 3.21SYY611 pKa = 11.88SDD613 pKa = 3.97EE614 pKa = 4.37ILIQANLVSSEE625 pKa = 4.32PDD627 pKa = 3.08LGAQNPDD634 pKa = 2.94ILVTEE639 pKa = 4.3AVVFLDD645 pKa = 5.53DD646 pKa = 5.33GGDD649 pKa = 3.76DD650 pKa = 3.94TGDD653 pKa = 3.38ASNTNPIAPQPADD666 pKa = 3.53SGSADD671 pKa = 3.45VMQHH675 pKa = 5.04MLGG678 pKa = 3.58
Molecular weight: 71.71 kDa
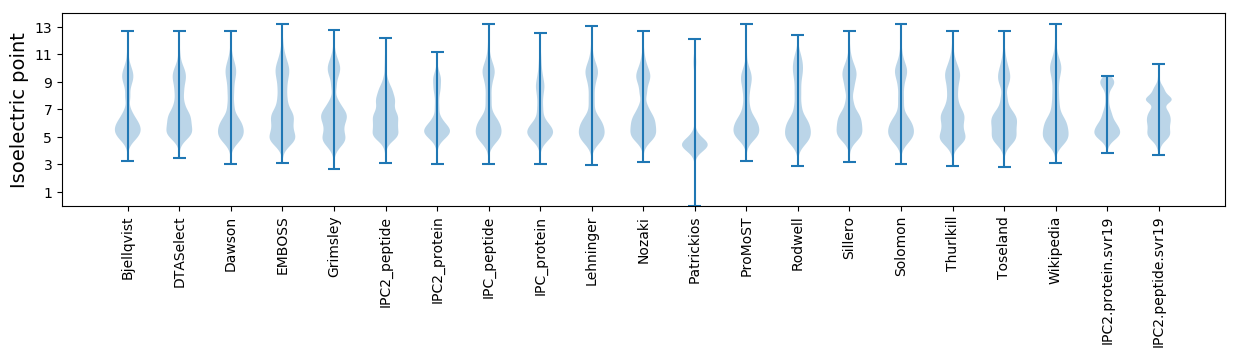
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q6LAC6|A0A0Q6LAC6_9RHIZ Amidase OS=Aminobacter sp. DSM 101952 OX=2735891 GN=ASC75_12330 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.11GGRR28 pKa = 11.84GVVAARR34 pKa = 11.84RR35 pKa = 11.84NRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.11GGRR28 pKa = 11.84GVVAARR34 pKa = 11.84RR35 pKa = 11.84NRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1488852 |
29 |
3861 |
306.9 |
33.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.44 ± 0.052 | 0.787 ± 0.012 |
5.761 ± 0.037 | 5.73 ± 0.033 |
3.877 ± 0.026 | 8.667 ± 0.038 |
2.002 ± 0.017 | 5.347 ± 0.025 |
3.651 ± 0.031 | 9.848 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.588 ± 0.015 | 2.672 ± 0.023 |
4.928 ± 0.025 | 3.009 ± 0.02 |
6.816 ± 0.039 | 5.536 ± 0.023 |
5.192 ± 0.029 | 7.621 ± 0.026 |
1.307 ± 0.018 | 2.221 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
