
Pseudoalteromonas sp. NBT06-2
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Pseudoalteromonadaceae; Pseudoalteromonas; unclassified Pseudoalteromonas
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
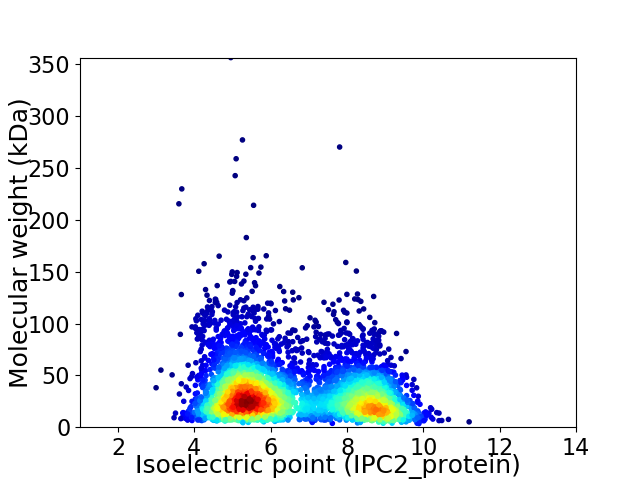
Virtual 2D-PAGE plot for 4921 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A269PT70|A0A269PT70_9GAMM Porin OS=Pseudoalteromonas sp. NBT06-2 OX=2025950 GN=CJF42_04650 PE=4 SV=1
MM1 pKa = 7.69KK2 pKa = 10.14FQKK5 pKa = 9.46TLLACTVSVFLMGCISDD22 pKa = 4.45NDD24 pKa = 4.27DD25 pKa = 3.55PSKK28 pKa = 11.19DD29 pKa = 3.87LIVPEE34 pKa = 4.18PVSVKK39 pKa = 10.48LVVPAEE45 pKa = 4.23GKK47 pKa = 11.05SMVEE51 pKa = 4.03DD52 pKa = 5.13FPLGMYY58 pKa = 7.66VTYY61 pKa = 10.36PEE63 pKa = 4.21SEE65 pKa = 3.98QRR67 pKa = 11.84ASAITSIDD75 pKa = 3.75VNWPFVDD82 pKa = 4.05VFQSGVEE89 pKa = 3.82YY90 pKa = 10.53GYY92 pKa = 10.47IDD94 pKa = 4.22GDD96 pKa = 3.78SDD98 pKa = 3.52EE99 pKa = 5.54LFYY102 pKa = 11.29HH103 pKa = 6.37SVEE106 pKa = 4.15NTNDD110 pKa = 3.1IAKK113 pKa = 10.21NAIDD117 pKa = 5.08LEE119 pKa = 4.19RR120 pKa = 11.84QEE122 pKa = 5.69DD123 pKa = 3.87ILWYY127 pKa = 9.12VTGNNLLFRR136 pKa = 11.84YY137 pKa = 9.86DD138 pKa = 3.5RR139 pKa = 11.84AQDD142 pKa = 4.28KK143 pKa = 10.79LVTWQVTGNSVFTEE157 pKa = 4.01LAIDD161 pKa = 3.7EE162 pKa = 4.87KK163 pKa = 11.42GEE165 pKa = 4.05QQVWLYY171 pKa = 11.54DD172 pKa = 3.66KK173 pKa = 10.05VTHH176 pKa = 6.17QLVIFNAIDD185 pKa = 3.51EE186 pKa = 4.44SSQVIPLEE194 pKa = 4.03GDD196 pKa = 3.29MSVEE200 pKa = 3.97GLGISEE206 pKa = 4.88DD207 pKa = 3.68SFLILAKK214 pKa = 10.47DD215 pKa = 3.66VAQGVVLHH223 pKa = 6.04YY224 pKa = 10.85SLAGTTLAYY233 pKa = 10.45NDD235 pKa = 2.92SWYY238 pKa = 11.26LEE240 pKa = 4.28GFGAAQFNDD249 pKa = 3.53IGLMPDD255 pKa = 2.9GRR257 pKa = 11.84IAVSNDD263 pKa = 2.84DD264 pKa = 3.85TEE266 pKa = 4.68NNLVFVMDD274 pKa = 4.37KK275 pKa = 9.61MAQIGAGPIKK285 pKa = 10.83DD286 pKa = 3.45NGEE289 pKa = 3.83LEE291 pKa = 4.63KK292 pKa = 10.62ISEE295 pKa = 4.24HH296 pKa = 5.96TLDD299 pKa = 3.85EE300 pKa = 4.96VIIQPSGIWALTDD313 pKa = 4.04DD314 pKa = 4.6SWMMITDD321 pKa = 3.61QAEE324 pKa = 4.28MFALDD329 pKa = 3.44ANFNVTEE336 pKa = 4.17KK337 pKa = 11.02VDD339 pKa = 3.89IEE341 pKa = 4.18FDD343 pKa = 4.92SINCNQGCTEE353 pKa = 4.99AIVGGDD359 pKa = 3.4NEE361 pKa = 5.11FYY363 pKa = 11.25ALTDD367 pKa = 3.54GGLVGHH373 pKa = 5.99FTKK376 pKa = 10.44TDD378 pKa = 3.15EE379 pKa = 4.55SYY381 pKa = 10.48TLAKK385 pKa = 9.88EE386 pKa = 3.94YY387 pKa = 10.53QINVTNSEE395 pKa = 4.23GEE397 pKa = 3.95NYY399 pKa = 9.93RR400 pKa = 11.84YY401 pKa = 10.26SGLGYY406 pKa = 10.27DD407 pKa = 4.12VSAQEE412 pKa = 4.61YY413 pKa = 10.61YY414 pKa = 10.91LIPDD418 pKa = 3.89QNDD421 pKa = 2.86EE422 pKa = 4.35DD423 pKa = 4.16ATDD426 pKa = 3.72EE427 pKa = 5.21LIILNSDD434 pKa = 3.72FTLKK438 pKa = 10.15EE439 pKa = 4.08RR440 pKa = 11.84YY441 pKa = 9.3DD442 pKa = 3.16ISYY445 pKa = 10.87AGEE448 pKa = 3.95TDD450 pKa = 3.58GSIFEE455 pKa = 4.39YY456 pKa = 10.52DD457 pKa = 3.34AQGVQYY463 pKa = 9.73HH464 pKa = 5.16QNNVYY469 pKa = 10.29VLSEE473 pKa = 4.45KK474 pKa = 9.31FTKK477 pKa = 10.59LIKK480 pKa = 10.62LNLVGEE486 pKa = 4.65IIAVYY491 pKa = 10.44DD492 pKa = 4.27LGHH495 pKa = 7.05EE496 pKa = 4.24DD497 pKa = 4.04VKK499 pKa = 11.35DD500 pKa = 3.8PSDD503 pKa = 3.3IALKK507 pKa = 10.63DD508 pKa = 3.37GQVYY512 pKa = 10.5VLGDD516 pKa = 3.79HH517 pKa = 6.89EE518 pKa = 4.95NDD520 pKa = 3.42EE521 pKa = 4.66PVPPLSVFDD530 pKa = 3.63IVEE533 pKa = 4.12YY534 pKa = 10.44EE535 pKa = 3.82
MM1 pKa = 7.69KK2 pKa = 10.14FQKK5 pKa = 9.46TLLACTVSVFLMGCISDD22 pKa = 4.45NDD24 pKa = 4.27DD25 pKa = 3.55PSKK28 pKa = 11.19DD29 pKa = 3.87LIVPEE34 pKa = 4.18PVSVKK39 pKa = 10.48LVVPAEE45 pKa = 4.23GKK47 pKa = 11.05SMVEE51 pKa = 4.03DD52 pKa = 5.13FPLGMYY58 pKa = 7.66VTYY61 pKa = 10.36PEE63 pKa = 4.21SEE65 pKa = 3.98QRR67 pKa = 11.84ASAITSIDD75 pKa = 3.75VNWPFVDD82 pKa = 4.05VFQSGVEE89 pKa = 3.82YY90 pKa = 10.53GYY92 pKa = 10.47IDD94 pKa = 4.22GDD96 pKa = 3.78SDD98 pKa = 3.52EE99 pKa = 5.54LFYY102 pKa = 11.29HH103 pKa = 6.37SVEE106 pKa = 4.15NTNDD110 pKa = 3.1IAKK113 pKa = 10.21NAIDD117 pKa = 5.08LEE119 pKa = 4.19RR120 pKa = 11.84QEE122 pKa = 5.69DD123 pKa = 3.87ILWYY127 pKa = 9.12VTGNNLLFRR136 pKa = 11.84YY137 pKa = 9.86DD138 pKa = 3.5RR139 pKa = 11.84AQDD142 pKa = 4.28KK143 pKa = 10.79LVTWQVTGNSVFTEE157 pKa = 4.01LAIDD161 pKa = 3.7EE162 pKa = 4.87KK163 pKa = 11.42GEE165 pKa = 4.05QQVWLYY171 pKa = 11.54DD172 pKa = 3.66KK173 pKa = 10.05VTHH176 pKa = 6.17QLVIFNAIDD185 pKa = 3.51EE186 pKa = 4.44SSQVIPLEE194 pKa = 4.03GDD196 pKa = 3.29MSVEE200 pKa = 3.97GLGISEE206 pKa = 4.88DD207 pKa = 3.68SFLILAKK214 pKa = 10.47DD215 pKa = 3.66VAQGVVLHH223 pKa = 6.04YY224 pKa = 10.85SLAGTTLAYY233 pKa = 10.45NDD235 pKa = 2.92SWYY238 pKa = 11.26LEE240 pKa = 4.28GFGAAQFNDD249 pKa = 3.53IGLMPDD255 pKa = 2.9GRR257 pKa = 11.84IAVSNDD263 pKa = 2.84DD264 pKa = 3.85TEE266 pKa = 4.68NNLVFVMDD274 pKa = 4.37KK275 pKa = 9.61MAQIGAGPIKK285 pKa = 10.83DD286 pKa = 3.45NGEE289 pKa = 3.83LEE291 pKa = 4.63KK292 pKa = 10.62ISEE295 pKa = 4.24HH296 pKa = 5.96TLDD299 pKa = 3.85EE300 pKa = 4.96VIIQPSGIWALTDD313 pKa = 4.04DD314 pKa = 4.6SWMMITDD321 pKa = 3.61QAEE324 pKa = 4.28MFALDD329 pKa = 3.44ANFNVTEE336 pKa = 4.17KK337 pKa = 11.02VDD339 pKa = 3.89IEE341 pKa = 4.18FDD343 pKa = 4.92SINCNQGCTEE353 pKa = 4.99AIVGGDD359 pKa = 3.4NEE361 pKa = 5.11FYY363 pKa = 11.25ALTDD367 pKa = 3.54GGLVGHH373 pKa = 5.99FTKK376 pKa = 10.44TDD378 pKa = 3.15EE379 pKa = 4.55SYY381 pKa = 10.48TLAKK385 pKa = 9.88EE386 pKa = 3.94YY387 pKa = 10.53QINVTNSEE395 pKa = 4.23GEE397 pKa = 3.95NYY399 pKa = 9.93RR400 pKa = 11.84YY401 pKa = 10.26SGLGYY406 pKa = 10.27DD407 pKa = 4.12VSAQEE412 pKa = 4.61YY413 pKa = 10.61YY414 pKa = 10.91LIPDD418 pKa = 3.89QNDD421 pKa = 2.86EE422 pKa = 4.35DD423 pKa = 4.16ATDD426 pKa = 3.72EE427 pKa = 5.21LIILNSDD434 pKa = 3.72FTLKK438 pKa = 10.15EE439 pKa = 4.08RR440 pKa = 11.84YY441 pKa = 9.3DD442 pKa = 3.16ISYY445 pKa = 10.87AGEE448 pKa = 3.95TDD450 pKa = 3.58GSIFEE455 pKa = 4.39YY456 pKa = 10.52DD457 pKa = 3.34AQGVQYY463 pKa = 9.73HH464 pKa = 5.16QNNVYY469 pKa = 10.29VLSEE473 pKa = 4.45KK474 pKa = 9.31FTKK477 pKa = 10.59LIKK480 pKa = 10.62LNLVGEE486 pKa = 4.65IIAVYY491 pKa = 10.44DD492 pKa = 4.27LGHH495 pKa = 7.05EE496 pKa = 4.24DD497 pKa = 4.04VKK499 pKa = 11.35DD500 pKa = 3.8PSDD503 pKa = 3.3IALKK507 pKa = 10.63DD508 pKa = 3.37GQVYY512 pKa = 10.5VLGDD516 pKa = 3.79HH517 pKa = 6.89EE518 pKa = 4.95NDD520 pKa = 3.42EE521 pKa = 4.66PVPPLSVFDD530 pKa = 3.63IVEE533 pKa = 4.12YY534 pKa = 10.44EE535 pKa = 3.82
Molecular weight: 59.78 kDa
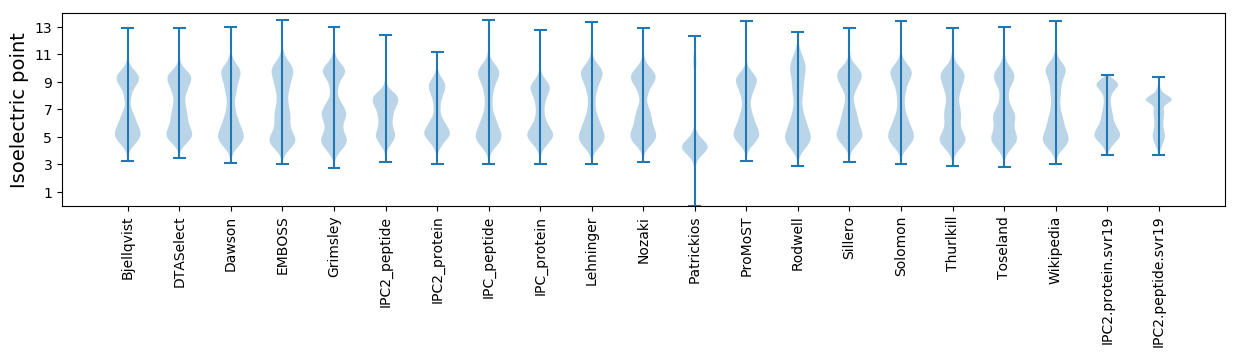
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A269PUH6|A0A269PUH6_9GAMM Bifunctional protein PutA OS=Pseudoalteromonas sp. NBT06-2 OX=2025950 GN=putA PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.36RR14 pKa = 11.84SHH16 pKa = 6.15GFRR19 pKa = 11.84ARR21 pKa = 11.84MATTNGRR28 pKa = 11.84NLINRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.79GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.36RR14 pKa = 11.84SHH16 pKa = 6.15GFRR19 pKa = 11.84ARR21 pKa = 11.84MATTNGRR28 pKa = 11.84NLINRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.79GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1573380 |
30 |
3199 |
319.7 |
35.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.298 ± 0.03 | 1.03 ± 0.015 |
5.523 ± 0.03 | 5.937 ± 0.029 |
4.506 ± 0.022 | 6.101 ± 0.036 |
2.143 ± 0.017 | 7.588 ± 0.029 |
6.824 ± 0.038 | 10.304 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.318 ± 0.018 | 5.615 ± 0.033 |
3.402 ± 0.019 | 4.454 ± 0.029 |
3.606 ± 0.026 | 7.172 ± 0.029 |
5.529 ± 0.028 | 6.168 ± 0.029 |
1.144 ± 0.014 | 3.339 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
