
Malassezia globosa (strain ATCC MYA-4612 / CBS 7966) (Dandruff-associated fungus)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Ustilaginomycotina; Malasseziomycetes; Malasseziales; Malasseziaceae; Malassezia; Malassezia globosa
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
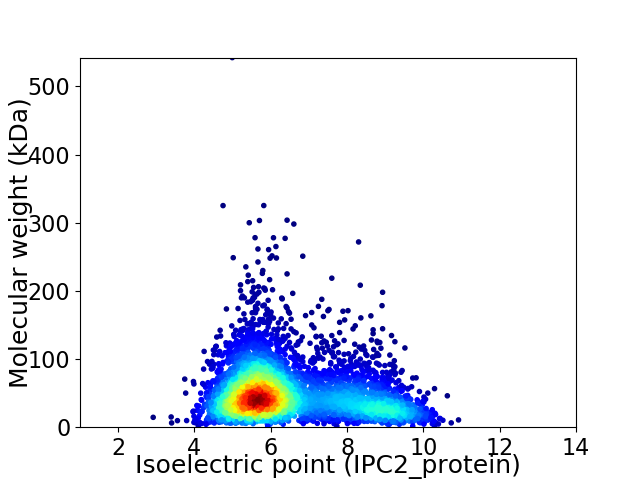
Virtual 2D-PAGE plot for 4274 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A8PY24|A8PY24_MALGO Glutaredoxin-like protein OS=Malassezia globosa (strain ATCC MYA-4612 / CBS 7966) OX=425265 GN=MGL_1576 PE=3 SV=1
MM1 pKa = 8.07DD2 pKa = 6.04LLDD5 pKa = 5.4AEE7 pKa = 4.45QQEE10 pKa = 4.74ALPFSLDD17 pKa = 3.64YY18 pKa = 10.67YY19 pKa = 11.12TDD21 pKa = 3.92AQDD24 pKa = 3.8LNYY27 pKa = 9.81MADD30 pKa = 4.01YY31 pKa = 10.9LHH33 pKa = 6.26EE34 pKa = 4.39TQPRR38 pKa = 11.84VARR41 pKa = 11.84MYY43 pKa = 10.4EE44 pKa = 3.83VLGDD48 pKa = 3.73IVQEE52 pKa = 3.94YY53 pKa = 10.44GLVSFEE59 pKa = 4.05TLAVEE64 pKa = 5.23EE65 pKa = 4.41KK66 pKa = 11.02ASMLHH71 pKa = 5.84LVEE74 pKa = 4.24VLDD77 pKa = 3.94RR78 pKa = 11.84AIGYY82 pKa = 7.27VARR85 pKa = 11.84DD86 pKa = 3.47SAA88 pKa = 4.09
MM1 pKa = 8.07DD2 pKa = 6.04LLDD5 pKa = 5.4AEE7 pKa = 4.45QQEE10 pKa = 4.74ALPFSLDD17 pKa = 3.64YY18 pKa = 10.67YY19 pKa = 11.12TDD21 pKa = 3.92AQDD24 pKa = 3.8LNYY27 pKa = 9.81MADD30 pKa = 4.01YY31 pKa = 10.9LHH33 pKa = 6.26EE34 pKa = 4.39TQPRR38 pKa = 11.84VARR41 pKa = 11.84MYY43 pKa = 10.4EE44 pKa = 3.83VLGDD48 pKa = 3.73IVQEE52 pKa = 3.94YY53 pKa = 10.44GLVSFEE59 pKa = 4.05TLAVEE64 pKa = 5.23EE65 pKa = 4.41KK66 pKa = 11.02ASMLHH71 pKa = 5.84LVEE74 pKa = 4.24VLDD77 pKa = 3.94RR78 pKa = 11.84AIGYY82 pKa = 7.27VARR85 pKa = 11.84DD86 pKa = 3.47SAA88 pKa = 4.09
Molecular weight: 10.06 kDa
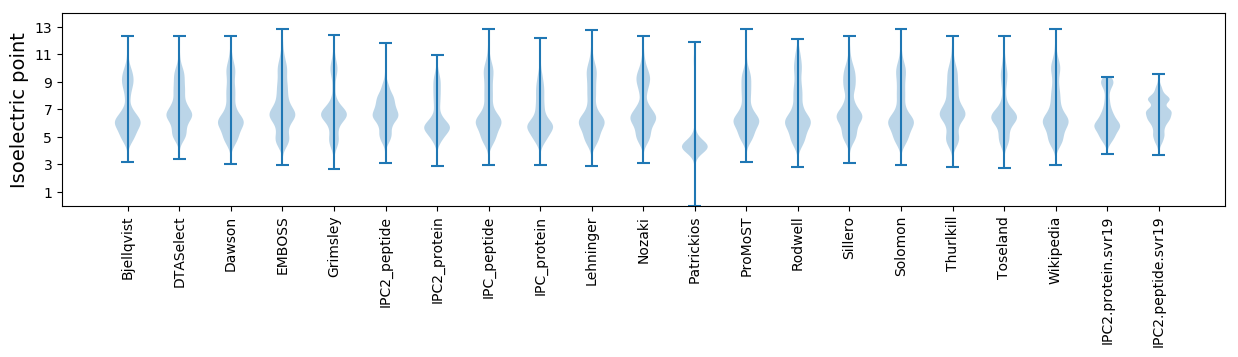
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A8Q258|A8Q258_MALGO Uncharacterized protein OS=Malassezia globosa (strain ATCC MYA-4612 / CBS 7966) OX=425265 GN=MGL_2143 PE=3 SV=1
MM1 pKa = 7.42MFQPSAVCKK10 pKa = 9.64TVVPDD15 pKa = 3.46TFRR18 pKa = 11.84VLRR21 pKa = 11.84SQRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84WINSTVHH33 pKa = 6.36NLFEE37 pKa = 5.35LIQVRR42 pKa = 11.84DD43 pKa = 3.64LCGTFCFSMRR53 pKa = 11.84FVVFMDD59 pKa = 3.76LVGTMVLPAAIAFTVFVVVQAILTPFQNKK88 pKa = 9.68GKK90 pKa = 10.22DD91 pKa = 3.37DD92 pKa = 4.04GEE94 pKa = 4.2KK95 pKa = 10.58KK96 pKa = 10.08EE97 pKa = 4.35FPTMPLVLLALILGLPGVLIVITSRR122 pKa = 11.84RR123 pKa = 11.84FMYY126 pKa = 8.67VIWMLVYY133 pKa = 10.76LISLPIWNLILPAYY147 pKa = 9.65SYY149 pKa = 9.94WHH151 pKa = 6.69MDD153 pKa = 3.41DD154 pKa = 4.78FSWGATRR161 pKa = 11.84MVQGEE166 pKa = 4.5KK167 pKa = 10.14KK168 pKa = 10.62GEE170 pKa = 4.09SHH172 pKa = 6.85GSAEE176 pKa = 4.37GEE178 pKa = 3.85FDD180 pKa = 4.24ASDD183 pKa = 3.15IVMKK187 pKa = 10.49RR188 pKa = 11.84YY189 pKa = 9.89VEE191 pKa = 4.14YY192 pKa = 10.47EE193 pKa = 3.56RR194 pKa = 11.84EE195 pKa = 4.05RR196 pKa = 11.84RR197 pKa = 11.84WRR199 pKa = 11.84SGMEE203 pKa = 3.87SRR205 pKa = 11.84DD206 pKa = 3.69APSEE210 pKa = 4.15SGGMSTLGSLNEE222 pKa = 4.25EE223 pKa = 4.38KK224 pKa = 10.76ALPPPGADD232 pKa = 2.82EE233 pKa = 3.98RR234 pKa = 11.84SARR237 pKa = 11.84HH238 pKa = 5.91RR239 pKa = 11.84LDD241 pKa = 3.39SVPLLEE247 pKa = 5.34LPAPLGSDD255 pKa = 2.92SRR257 pKa = 11.84QRR259 pKa = 11.84AGTMSPPVPGGKK271 pKa = 9.92GPNGARR277 pKa = 11.84SSPAGTTSPPNPWTSSGALTQTGPAPLTAQRR308 pKa = 11.84VNPSPNPSASAMARR322 pKa = 11.84QHH324 pKa = 6.61LFGATPTSAAHH335 pKa = 6.62ASMPVRR341 pKa = 11.84PPGTISPPPRR351 pKa = 11.84PSNIGASGPVSPPSFRR367 pKa = 11.84GNVSPPPMFPAGPLSPVNTRR387 pKa = 11.84PTPPVRR393 pKa = 11.84PVAPSPSGWNAASQPARR410 pKa = 11.84PANTQAYY417 pKa = 7.09PSPPSSGTQPGSAPTLAGFQPLSPPSASRR446 pKa = 11.84PPPLRR451 pKa = 11.84APSNAQQMRR460 pKa = 11.84GDD462 pKa = 3.59VRR464 pKa = 11.84RR465 pKa = 11.84VSLVDD470 pKa = 4.24DD471 pKa = 4.79GPVATSGGSMRR482 pKa = 11.84QVVRR486 pKa = 11.84GARR489 pKa = 11.84RR490 pKa = 11.84QSSINSGAPNSASTASPPSNSNPFSRR516 pKa = 11.84PANPFSPPPKK526 pKa = 10.17
MM1 pKa = 7.42MFQPSAVCKK10 pKa = 9.64TVVPDD15 pKa = 3.46TFRR18 pKa = 11.84VLRR21 pKa = 11.84SQRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84WINSTVHH33 pKa = 6.36NLFEE37 pKa = 5.35LIQVRR42 pKa = 11.84DD43 pKa = 3.64LCGTFCFSMRR53 pKa = 11.84FVVFMDD59 pKa = 3.76LVGTMVLPAAIAFTVFVVVQAILTPFQNKK88 pKa = 9.68GKK90 pKa = 10.22DD91 pKa = 3.37DD92 pKa = 4.04GEE94 pKa = 4.2KK95 pKa = 10.58KK96 pKa = 10.08EE97 pKa = 4.35FPTMPLVLLALILGLPGVLIVITSRR122 pKa = 11.84RR123 pKa = 11.84FMYY126 pKa = 8.67VIWMLVYY133 pKa = 10.76LISLPIWNLILPAYY147 pKa = 9.65SYY149 pKa = 9.94WHH151 pKa = 6.69MDD153 pKa = 3.41DD154 pKa = 4.78FSWGATRR161 pKa = 11.84MVQGEE166 pKa = 4.5KK167 pKa = 10.14KK168 pKa = 10.62GEE170 pKa = 4.09SHH172 pKa = 6.85GSAEE176 pKa = 4.37GEE178 pKa = 3.85FDD180 pKa = 4.24ASDD183 pKa = 3.15IVMKK187 pKa = 10.49RR188 pKa = 11.84YY189 pKa = 9.89VEE191 pKa = 4.14YY192 pKa = 10.47EE193 pKa = 3.56RR194 pKa = 11.84EE195 pKa = 4.05RR196 pKa = 11.84RR197 pKa = 11.84WRR199 pKa = 11.84SGMEE203 pKa = 3.87SRR205 pKa = 11.84DD206 pKa = 3.69APSEE210 pKa = 4.15SGGMSTLGSLNEE222 pKa = 4.25EE223 pKa = 4.38KK224 pKa = 10.76ALPPPGADD232 pKa = 2.82EE233 pKa = 3.98RR234 pKa = 11.84SARR237 pKa = 11.84HH238 pKa = 5.91RR239 pKa = 11.84LDD241 pKa = 3.39SVPLLEE247 pKa = 5.34LPAPLGSDD255 pKa = 2.92SRR257 pKa = 11.84QRR259 pKa = 11.84AGTMSPPVPGGKK271 pKa = 9.92GPNGARR277 pKa = 11.84SSPAGTTSPPNPWTSSGALTQTGPAPLTAQRR308 pKa = 11.84VNPSPNPSASAMARR322 pKa = 11.84QHH324 pKa = 6.61LFGATPTSAAHH335 pKa = 6.62ASMPVRR341 pKa = 11.84PPGTISPPPRR351 pKa = 11.84PSNIGASGPVSPPSFRR367 pKa = 11.84GNVSPPPMFPAGPLSPVNTRR387 pKa = 11.84PTPPVRR393 pKa = 11.84PVAPSPSGWNAASQPARR410 pKa = 11.84PANTQAYY417 pKa = 7.09PSPPSSGTQPGSAPTLAGFQPLSPPSASRR446 pKa = 11.84PPPLRR451 pKa = 11.84APSNAQQMRR460 pKa = 11.84GDD462 pKa = 3.59VRR464 pKa = 11.84RR465 pKa = 11.84VSLVDD470 pKa = 4.24DD471 pKa = 4.79GPVATSGGSMRR482 pKa = 11.84QVVRR486 pKa = 11.84GARR489 pKa = 11.84RR490 pKa = 11.84QSSINSGAPNSASTASPPSNSNPFSRR516 pKa = 11.84PANPFSPPPKK526 pKa = 10.17
Molecular weight: 56.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
2064385 |
14 |
4917 |
483.0 |
53.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.527 ± 0.046 | 1.292 ± 0.012 |
5.843 ± 0.024 | 5.865 ± 0.034 |
3.245 ± 0.024 | 5.839 ± 0.033 |
3.16 ± 0.02 | 4.037 ± 0.026 |
3.877 ± 0.033 | 9.45 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.727 ± 0.013 | 3.037 ± 0.023 |
6.123 ± 0.045 | 4.27 ± 0.022 |
6.743 ± 0.032 | 8.674 ± 0.05 |
5.871 ± 0.023 | 6.526 ± 0.029 |
1.339 ± 0.013 | 2.553 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
