
Nitrosococcus oceani (strain ATCC 19707 / BCRC 17464 / NCIMB 11848 / C-107)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Chromatiales; Chromatiaceae; Nitrosococcus; Nitrosococcus oceani
Average proteome isoelectric point is 6.73
Get precalculated fractions of proteins
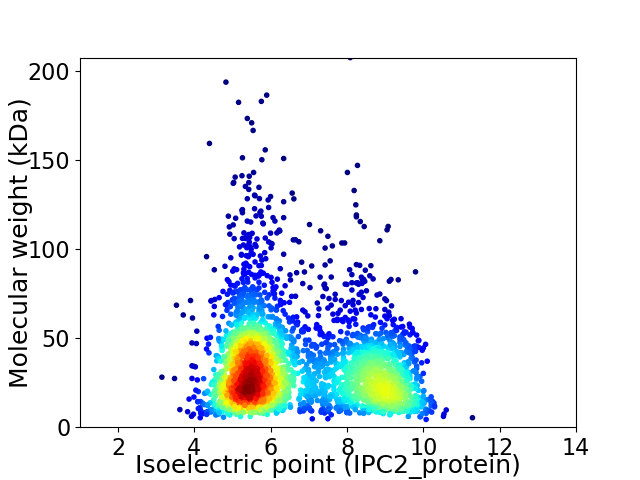
Virtual 2D-PAGE plot for 2995 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q3JEM2|Q3JEM2_NITOC Thiosulfate sulfurtransferase OS=Nitrosococcus oceani (strain ATCC 19707 / BCRC 17464 / NCIMB 11848 / C-107) OX=323261 GN=Noc_0191 PE=4 SV=1
MM1 pKa = 7.51KK2 pKa = 10.34NLARR6 pKa = 11.84HH7 pKa = 5.75CFIFLGLLLVPTLGAAGLNIFPEE30 pKa = 4.68LDD32 pKa = 3.58SNDD35 pKa = 3.85NNNTSSNNTSQEE47 pKa = 3.88STQNFSNTADD57 pKa = 4.2PEE59 pKa = 4.4PEE61 pKa = 4.31ADD63 pKa = 3.79TNASPPANKK72 pKa = 10.22DD73 pKa = 3.3EE74 pKa = 5.32IIAEE78 pKa = 4.39TKK80 pKa = 10.32AGCATDD86 pKa = 3.84PASCGVTLSSFLDD99 pKa = 3.35STGFGEE105 pKa = 4.77TEE107 pKa = 4.25PNNHH111 pKa = 6.15AMGADD116 pKa = 3.0AMEE119 pKa = 4.75FGVEE123 pKa = 4.03YY124 pKa = 10.53AGQLYY129 pKa = 10.53SPEE132 pKa = 4.09DD133 pKa = 3.71VDD135 pKa = 3.39WFRR138 pKa = 11.84ITTTEE143 pKa = 4.09SNQMLTVNFNVPGLNDD159 pKa = 3.07ITGWNLSIRR168 pKa = 11.84DD169 pKa = 3.44SGGNIFSEE177 pKa = 5.26VYY179 pKa = 9.59TGFDD183 pKa = 4.06FGPEE187 pKa = 4.11SPLQTILSRR196 pKa = 11.84AGTYY200 pKa = 9.52YY201 pKa = 11.08VVVKK205 pKa = 10.11SLKK208 pKa = 9.24QAQEE212 pKa = 3.97EE213 pKa = 4.6TRR215 pKa = 11.84SSDD218 pKa = 3.35QSGEE222 pKa = 3.58ADD224 pKa = 4.93LIYY227 pKa = 10.73EE228 pKa = 4.52HH229 pKa = 7.05LPHH232 pKa = 7.41EE233 pKa = 4.39YY234 pKa = 10.23RR235 pKa = 11.84LAAFLGDD242 pKa = 3.66SQVTTEE248 pKa = 4.23PLDD251 pKa = 3.66VNFFDD256 pKa = 6.29AEE258 pKa = 4.26VEE260 pKa = 4.36PNDD263 pKa = 4.81SRR265 pKa = 11.84DD266 pKa = 3.38EE267 pKa = 4.41ANPLTFATPLASNVTMEE284 pKa = 4.44GLISGPLIFGSVGFAFEE301 pKa = 4.49EE302 pKa = 4.33DD303 pKa = 3.1WFVYY307 pKa = 8.93DD308 pKa = 3.52TAGNEE313 pKa = 3.7ILSIEE318 pKa = 4.22FCASQDD324 pKa = 4.1CEE326 pKa = 4.15DD327 pKa = 4.1STWQVTVYY335 pKa = 10.97DD336 pKa = 3.97EE337 pKa = 4.32NEE339 pKa = 3.88RR340 pKa = 11.84MLLTGRR346 pKa = 11.84TDD348 pKa = 3.31MEE350 pKa = 3.93QNYY353 pKa = 9.95YY354 pKa = 10.72LGIRR358 pKa = 11.84NPGKK362 pKa = 8.64YY363 pKa = 10.13FIRR366 pKa = 11.84IGVAPALDD374 pKa = 4.12EE375 pKa = 4.42EE376 pKa = 4.91SGGAQYY382 pKa = 11.08VCSIDD387 pKa = 3.15PTMPLKK393 pKa = 10.69DD394 pKa = 3.83CPSPSEE400 pKa = 3.79RR401 pKa = 11.84TLLVEE406 pKa = 5.02SPWHH410 pKa = 5.52QYY412 pKa = 10.38NFTVTSTKK420 pKa = 10.41LPPLMSEE427 pKa = 3.99VDD429 pKa = 3.57NPP431 pKa = 3.65
MM1 pKa = 7.51KK2 pKa = 10.34NLARR6 pKa = 11.84HH7 pKa = 5.75CFIFLGLLLVPTLGAAGLNIFPEE30 pKa = 4.68LDD32 pKa = 3.58SNDD35 pKa = 3.85NNNTSSNNTSQEE47 pKa = 3.88STQNFSNTADD57 pKa = 4.2PEE59 pKa = 4.4PEE61 pKa = 4.31ADD63 pKa = 3.79TNASPPANKK72 pKa = 10.22DD73 pKa = 3.3EE74 pKa = 5.32IIAEE78 pKa = 4.39TKK80 pKa = 10.32AGCATDD86 pKa = 3.84PASCGVTLSSFLDD99 pKa = 3.35STGFGEE105 pKa = 4.77TEE107 pKa = 4.25PNNHH111 pKa = 6.15AMGADD116 pKa = 3.0AMEE119 pKa = 4.75FGVEE123 pKa = 4.03YY124 pKa = 10.53AGQLYY129 pKa = 10.53SPEE132 pKa = 4.09DD133 pKa = 3.71VDD135 pKa = 3.39WFRR138 pKa = 11.84ITTTEE143 pKa = 4.09SNQMLTVNFNVPGLNDD159 pKa = 3.07ITGWNLSIRR168 pKa = 11.84DD169 pKa = 3.44SGGNIFSEE177 pKa = 5.26VYY179 pKa = 9.59TGFDD183 pKa = 4.06FGPEE187 pKa = 4.11SPLQTILSRR196 pKa = 11.84AGTYY200 pKa = 9.52YY201 pKa = 11.08VVVKK205 pKa = 10.11SLKK208 pKa = 9.24QAQEE212 pKa = 3.97EE213 pKa = 4.6TRR215 pKa = 11.84SSDD218 pKa = 3.35QSGEE222 pKa = 3.58ADD224 pKa = 4.93LIYY227 pKa = 10.73EE228 pKa = 4.52HH229 pKa = 7.05LPHH232 pKa = 7.41EE233 pKa = 4.39YY234 pKa = 10.23RR235 pKa = 11.84LAAFLGDD242 pKa = 3.66SQVTTEE248 pKa = 4.23PLDD251 pKa = 3.66VNFFDD256 pKa = 6.29AEE258 pKa = 4.26VEE260 pKa = 4.36PNDD263 pKa = 4.81SRR265 pKa = 11.84DD266 pKa = 3.38EE267 pKa = 4.41ANPLTFATPLASNVTMEE284 pKa = 4.44GLISGPLIFGSVGFAFEE301 pKa = 4.49EE302 pKa = 4.33DD303 pKa = 3.1WFVYY307 pKa = 8.93DD308 pKa = 3.52TAGNEE313 pKa = 3.7ILSIEE318 pKa = 4.22FCASQDD324 pKa = 4.1CEE326 pKa = 4.15DD327 pKa = 4.1STWQVTVYY335 pKa = 10.97DD336 pKa = 3.97EE337 pKa = 4.32NEE339 pKa = 3.88RR340 pKa = 11.84MLLTGRR346 pKa = 11.84TDD348 pKa = 3.31MEE350 pKa = 3.93QNYY353 pKa = 9.95YY354 pKa = 10.72LGIRR358 pKa = 11.84NPGKK362 pKa = 8.64YY363 pKa = 10.13FIRR366 pKa = 11.84IGVAPALDD374 pKa = 4.12EE375 pKa = 4.42EE376 pKa = 4.91SGGAQYY382 pKa = 11.08VCSIDD387 pKa = 3.15PTMPLKK393 pKa = 10.69DD394 pKa = 3.83CPSPSEE400 pKa = 3.79RR401 pKa = 11.84TLLVEE406 pKa = 5.02SPWHH410 pKa = 5.52QYY412 pKa = 10.38NFTVTSTKK420 pKa = 10.41LPPLMSEE427 pKa = 3.99VDD429 pKa = 3.57NPP431 pKa = 3.65
Molecular weight: 47.27 kDa
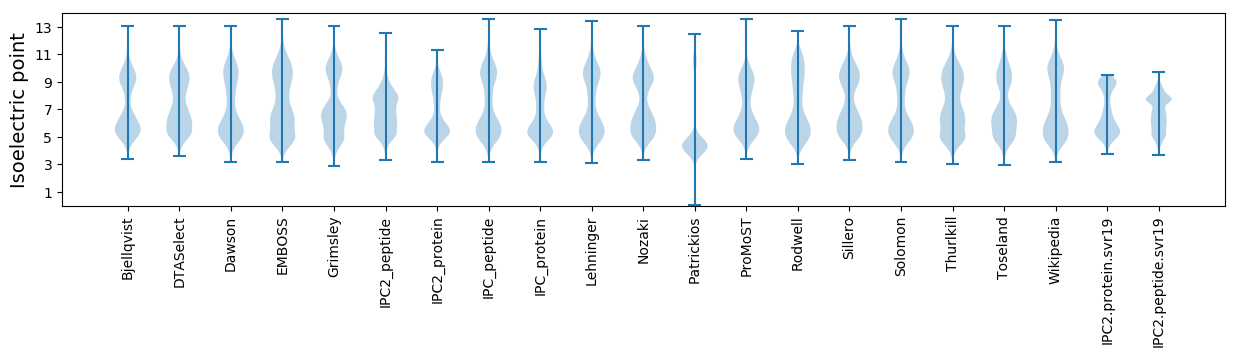
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q3J6M5|Q3J6M5_NITOC ATP synthase subunit a OS=Nitrosococcus oceani (strain ATCC 19707 / BCRC 17464 / NCIMB 11848 / C-107) OX=323261 GN=atpB PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPKK8 pKa = 8.02NLKK11 pKa = 9.99RR12 pKa = 11.84KK13 pKa = 7.29RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MRR23 pKa = 11.84TQGGRR28 pKa = 11.84AIINRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.73GRR39 pKa = 11.84KK40 pKa = 9.04RR41 pKa = 11.84LTAA44 pKa = 4.18
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPKK8 pKa = 8.02NLKK11 pKa = 9.99RR12 pKa = 11.84KK13 pKa = 7.29RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MRR23 pKa = 11.84TQGGRR28 pKa = 11.84AIINRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.73GRR39 pKa = 11.84KK40 pKa = 9.04RR41 pKa = 11.84LTAA44 pKa = 4.18
Molecular weight: 5.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
987868 |
37 |
1867 |
329.8 |
36.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.483 ± 0.051 | 0.996 ± 0.017 |
4.707 ± 0.031 | 6.604 ± 0.041 |
3.945 ± 0.03 | 7.61 ± 0.039 |
2.419 ± 0.022 | 5.859 ± 0.03 |
4.182 ± 0.038 | 11.286 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.165 ± 0.021 | 3.183 ± 0.023 |
4.961 ± 0.036 | 4.472 ± 0.033 |
6.583 ± 0.041 | 5.649 ± 0.034 |
4.969 ± 0.031 | 6.573 ± 0.035 |
1.484 ± 0.022 | 2.87 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
