
Legionella oakridgensis ATCC 33761 = DSM 21215
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Legionellales; Legionellaceae; Legionella; Legionella oakridgensis
Average proteome isoelectric point is 6.87
Get precalculated fractions of proteins
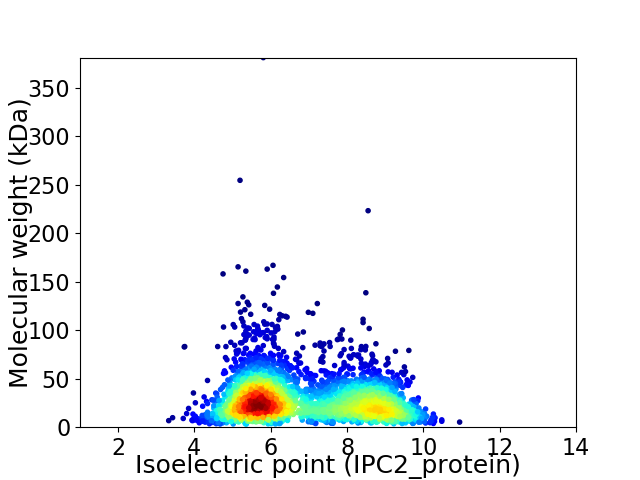
Virtual 2D-PAGE plot for 2907 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W0BHA3|W0BHA3_9GAMM Coenzyme F420-reducing hydrogenase alpha subunit OS=Legionella oakridgensis ATCC 33761 = DSM 21215 OX=1268635 GN=Loa_02480 PE=3 SV=1
MM1 pKa = 7.49SNSDD5 pKa = 3.62FEE7 pKa = 4.55QFLTALGAFEE17 pKa = 5.21SGIDD21 pKa = 3.5TTQTQTTSWMQYY33 pKa = 10.18LQVFDD38 pKa = 4.77PNRR41 pKa = 11.84GNVDD45 pKa = 3.65PNSVNLSDD53 pKa = 4.62PNDD56 pKa = 3.79LAEE59 pKa = 4.5LQYY62 pKa = 10.72HH63 pKa = 5.06VHH65 pKa = 5.66NTLGFLGKK73 pKa = 10.09YY74 pKa = 8.49QFGEE78 pKa = 4.07PLLIDD83 pKa = 4.07LGYY86 pKa = 10.69YY87 pKa = 10.18SPAPTGYY94 pKa = 10.76YY95 pKa = 8.46GTTATNEE102 pKa = 4.0WQGTWTGKK110 pKa = 10.41NGVDD114 pKa = 3.44SKK116 pKa = 11.6EE117 pKa = 3.82EE118 pKa = 3.77FMTNVQEE125 pKa = 4.04LAIRR129 pKa = 11.84EE130 pKa = 4.21AFAMNMGVINQYY142 pKa = 10.96LNQAGKK148 pKa = 8.9TIDD151 pKa = 4.36DD152 pKa = 4.01FLGHH156 pKa = 6.76EE157 pKa = 4.32FTYY160 pKa = 10.69TRR162 pKa = 11.84FGEE165 pKa = 3.99AHH167 pKa = 6.79IATVTMSGILASAHH181 pKa = 5.41LQGPGGVAQLLLHH194 pKa = 6.72NIASSDD200 pKa = 3.6EE201 pKa = 4.18YY202 pKa = 10.41GTNILFYY209 pKa = 8.68MDD211 pKa = 4.15KK212 pKa = 10.59FGGYY216 pKa = 7.47HH217 pKa = 5.8TPFGTNGDD225 pKa = 4.06DD226 pKa = 3.82FLVGSDD232 pKa = 3.73YY233 pKa = 11.6SEE235 pKa = 4.01TFMGEE240 pKa = 4.0GGHH243 pKa = 6.31NEE245 pKa = 4.09YY246 pKa = 9.04EE247 pKa = 4.3TGGGNDD253 pKa = 4.6KK254 pKa = 10.79IIIAQNITGNDD265 pKa = 3.52VIQDD269 pKa = 3.11FDD271 pKa = 3.33IARR274 pKa = 11.84DD275 pKa = 3.92VISLTKK281 pKa = 10.54FPGLTFADD289 pKa = 4.48LSITALNNNAVIHH302 pKa = 6.16FPNGQTVTLNNVDD315 pKa = 3.62ASQISANQFVSGPYY329 pKa = 9.97KK330 pKa = 10.64IMWNAGSGDD339 pKa = 3.8TIIEE343 pKa = 4.21NFNLQHH349 pKa = 7.28DD350 pKa = 5.44LIDD353 pKa = 4.08LNYY356 pKa = 10.93AFASNNLALYY366 pKa = 9.05EE367 pKa = 4.31EE368 pKa = 4.87NGSAVIDD375 pKa = 4.03VIGNNQRR382 pKa = 11.84IILAGIEE389 pKa = 4.2LDD391 pKa = 3.71EE392 pKa = 4.89LSPFHH397 pKa = 7.16FIKK400 pKa = 10.8APTDD404 pKa = 3.7FAQVHH409 pKa = 5.96FGINSDD415 pKa = 3.45HH416 pKa = 6.7TPPVVDD422 pKa = 4.76PGDD425 pKa = 3.98ANTGNDD431 pKa = 3.26NPADD435 pKa = 3.9GGDD438 pKa = 3.6NNNQQGTMDD447 pKa = 4.32GDD449 pKa = 3.68VFAYY453 pKa = 7.38TWNWGARR460 pKa = 11.84DD461 pKa = 3.9VIQGFDD467 pKa = 3.28ATEE470 pKa = 4.07DD471 pKa = 4.06KK472 pKa = 10.8IDD474 pKa = 4.21LQNFWTSFDD483 pKa = 4.26RR484 pKa = 11.84IQFYY488 pKa = 11.47DD489 pKa = 3.59NANGDD494 pKa = 3.88AVIDD498 pKa = 3.82LTTLNNQTITLADD511 pKa = 3.42VSVQQLSSANFIGVTGTMGIHH532 pKa = 6.24NEE534 pKa = 4.16PNNPPANEE542 pKa = 4.11PTDD545 pKa = 3.83PSPPSEE551 pKa = 4.16NDD553 pKa = 3.53AIPPEE558 pKa = 4.48DD559 pKa = 3.59TTGSGDD565 pKa = 3.53SYY567 pKa = 11.21VFTWKK572 pKa = 9.52WGAAEE577 pKa = 4.37TINNFDD583 pKa = 3.31VGQDD587 pKa = 3.74VIDD590 pKa = 5.08LKK592 pKa = 11.12SFWSSYY598 pKa = 7.87SQFSIYY604 pKa = 10.73DD605 pKa = 3.63NPQGNAVIDD614 pKa = 4.13LSQLNNQTITIQGVSSAEE632 pKa = 3.73LSAKK636 pKa = 10.47NIVGVAGTIDD646 pKa = 3.31QALTGHH652 pKa = 6.99SIPPSDD658 pKa = 3.96HH659 pKa = 6.96TDD661 pKa = 3.52PPDD664 pKa = 3.38TGTVTDD670 pKa = 4.91VISHH674 pKa = 6.87PVNSTNLVASDD685 pKa = 3.76AVQDD689 pKa = 3.72VFNFTWSWGSQNVIDD704 pKa = 5.15GFNPTQDD711 pKa = 3.38TVDD714 pKa = 4.27LSRR717 pKa = 11.84FWTTNDD723 pKa = 3.72QVDD726 pKa = 4.25IYY728 pKa = 11.79NNAQGDD734 pKa = 4.09AVIDD738 pKa = 3.99LSALNNQTITLLGVNADD755 pKa = 5.46DD756 pKa = 5.2LNQDD760 pKa = 3.08NVIFF764 pKa = 4.45
MM1 pKa = 7.49SNSDD5 pKa = 3.62FEE7 pKa = 4.55QFLTALGAFEE17 pKa = 5.21SGIDD21 pKa = 3.5TTQTQTTSWMQYY33 pKa = 10.18LQVFDD38 pKa = 4.77PNRR41 pKa = 11.84GNVDD45 pKa = 3.65PNSVNLSDD53 pKa = 4.62PNDD56 pKa = 3.79LAEE59 pKa = 4.5LQYY62 pKa = 10.72HH63 pKa = 5.06VHH65 pKa = 5.66NTLGFLGKK73 pKa = 10.09YY74 pKa = 8.49QFGEE78 pKa = 4.07PLLIDD83 pKa = 4.07LGYY86 pKa = 10.69YY87 pKa = 10.18SPAPTGYY94 pKa = 10.76YY95 pKa = 8.46GTTATNEE102 pKa = 4.0WQGTWTGKK110 pKa = 10.41NGVDD114 pKa = 3.44SKK116 pKa = 11.6EE117 pKa = 3.82EE118 pKa = 3.77FMTNVQEE125 pKa = 4.04LAIRR129 pKa = 11.84EE130 pKa = 4.21AFAMNMGVINQYY142 pKa = 10.96LNQAGKK148 pKa = 8.9TIDD151 pKa = 4.36DD152 pKa = 4.01FLGHH156 pKa = 6.76EE157 pKa = 4.32FTYY160 pKa = 10.69TRR162 pKa = 11.84FGEE165 pKa = 3.99AHH167 pKa = 6.79IATVTMSGILASAHH181 pKa = 5.41LQGPGGVAQLLLHH194 pKa = 6.72NIASSDD200 pKa = 3.6EE201 pKa = 4.18YY202 pKa = 10.41GTNILFYY209 pKa = 8.68MDD211 pKa = 4.15KK212 pKa = 10.59FGGYY216 pKa = 7.47HH217 pKa = 5.8TPFGTNGDD225 pKa = 4.06DD226 pKa = 3.82FLVGSDD232 pKa = 3.73YY233 pKa = 11.6SEE235 pKa = 4.01TFMGEE240 pKa = 4.0GGHH243 pKa = 6.31NEE245 pKa = 4.09YY246 pKa = 9.04EE247 pKa = 4.3TGGGNDD253 pKa = 4.6KK254 pKa = 10.79IIIAQNITGNDD265 pKa = 3.52VIQDD269 pKa = 3.11FDD271 pKa = 3.33IARR274 pKa = 11.84DD275 pKa = 3.92VISLTKK281 pKa = 10.54FPGLTFADD289 pKa = 4.48LSITALNNNAVIHH302 pKa = 6.16FPNGQTVTLNNVDD315 pKa = 3.62ASQISANQFVSGPYY329 pKa = 9.97KK330 pKa = 10.64IMWNAGSGDD339 pKa = 3.8TIIEE343 pKa = 4.21NFNLQHH349 pKa = 7.28DD350 pKa = 5.44LIDD353 pKa = 4.08LNYY356 pKa = 10.93AFASNNLALYY366 pKa = 9.05EE367 pKa = 4.31EE368 pKa = 4.87NGSAVIDD375 pKa = 4.03VIGNNQRR382 pKa = 11.84IILAGIEE389 pKa = 4.2LDD391 pKa = 3.71EE392 pKa = 4.89LSPFHH397 pKa = 7.16FIKK400 pKa = 10.8APTDD404 pKa = 3.7FAQVHH409 pKa = 5.96FGINSDD415 pKa = 3.45HH416 pKa = 6.7TPPVVDD422 pKa = 4.76PGDD425 pKa = 3.98ANTGNDD431 pKa = 3.26NPADD435 pKa = 3.9GGDD438 pKa = 3.6NNNQQGTMDD447 pKa = 4.32GDD449 pKa = 3.68VFAYY453 pKa = 7.38TWNWGARR460 pKa = 11.84DD461 pKa = 3.9VIQGFDD467 pKa = 3.28ATEE470 pKa = 4.07DD471 pKa = 4.06KK472 pKa = 10.8IDD474 pKa = 4.21LQNFWTSFDD483 pKa = 4.26RR484 pKa = 11.84IQFYY488 pKa = 11.47DD489 pKa = 3.59NANGDD494 pKa = 3.88AVIDD498 pKa = 3.82LTTLNNQTITLADD511 pKa = 3.42VSVQQLSSANFIGVTGTMGIHH532 pKa = 6.24NEE534 pKa = 4.16PNNPPANEE542 pKa = 4.11PTDD545 pKa = 3.83PSPPSEE551 pKa = 4.16NDD553 pKa = 3.53AIPPEE558 pKa = 4.48DD559 pKa = 3.59TTGSGDD565 pKa = 3.53SYY567 pKa = 11.21VFTWKK572 pKa = 9.52WGAAEE577 pKa = 4.37TINNFDD583 pKa = 3.31VGQDD587 pKa = 3.74VIDD590 pKa = 5.08LKK592 pKa = 11.12SFWSSYY598 pKa = 7.87SQFSIYY604 pKa = 10.73DD605 pKa = 3.63NPQGNAVIDD614 pKa = 4.13LSQLNNQTITIQGVSSAEE632 pKa = 3.73LSAKK636 pKa = 10.47NIVGVAGTIDD646 pKa = 3.31QALTGHH652 pKa = 6.99SIPPSDD658 pKa = 3.96HH659 pKa = 6.96TDD661 pKa = 3.52PPDD664 pKa = 3.38TGTVTDD670 pKa = 4.91VISHH674 pKa = 6.87PVNSTNLVASDD685 pKa = 3.76AVQDD689 pKa = 3.72VFNFTWSWGSQNVIDD704 pKa = 5.15GFNPTQDD711 pKa = 3.38TVDD714 pKa = 4.27LSRR717 pKa = 11.84FWTTNDD723 pKa = 3.72QVDD726 pKa = 4.25IYY728 pKa = 11.79NNAQGDD734 pKa = 4.09AVIDD738 pKa = 3.99LSALNNQTITLLGVNADD755 pKa = 5.46DD756 pKa = 5.2LNQDD760 pKa = 3.08NVIFF764 pKa = 4.45
Molecular weight: 83.09 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W0BDB8|W0BDB8_9GAMM Histidinol-phosphate/aromatic aminotransferase and cobyric acid decarboxylase OS=Legionella oakridgensis ATCC 33761 = DSM 21215 OX=1268635 GN=Loa_01076 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.18RR12 pKa = 11.84KK13 pKa = 9.04RR14 pKa = 11.84DD15 pKa = 3.36HH16 pKa = 6.66GFRR19 pKa = 11.84ARR21 pKa = 11.84MATRR25 pKa = 11.84AGRR28 pKa = 11.84LVLKK32 pKa = 10.36RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.84GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.18RR12 pKa = 11.84KK13 pKa = 9.04RR14 pKa = 11.84DD15 pKa = 3.36HH16 pKa = 6.66GFRR19 pKa = 11.84ARR21 pKa = 11.84MATRR25 pKa = 11.84AGRR28 pKa = 11.84LVLKK32 pKa = 10.36RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.84GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
808281 |
29 |
3405 |
278.0 |
31.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.324 ± 0.048 | 1.236 ± 0.016 |
4.694 ± 0.033 | 5.863 ± 0.048 |
4.374 ± 0.033 | 6.178 ± 0.048 |
2.846 ± 0.022 | 7.237 ± 0.043 |
5.634 ± 0.04 | 11.051 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.621 ± 0.023 | 4.263 ± 0.037 |
4.15 ± 0.026 | 4.787 ± 0.033 |
4.727 ± 0.036 | 6.15 ± 0.038 |
5.21 ± 0.033 | 6.058 ± 0.042 |
1.176 ± 0.016 | 3.421 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
