
Burkholderia plantarii
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Burkholderiaceae; Burkholderia
Average proteome isoelectric point is 6.85
Get precalculated fractions of proteins
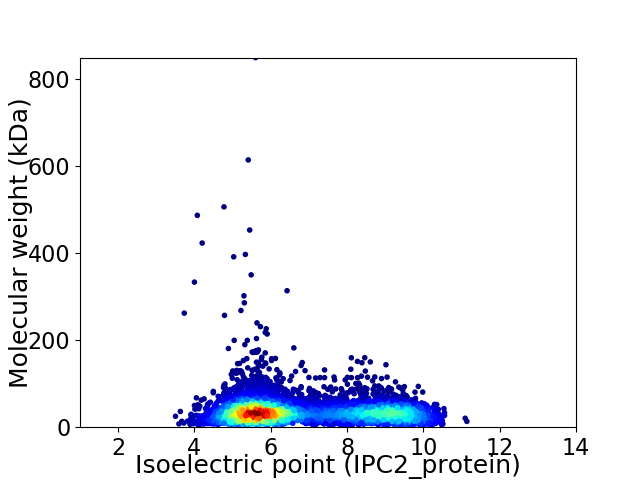
Virtual 2D-PAGE plot for 6432 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
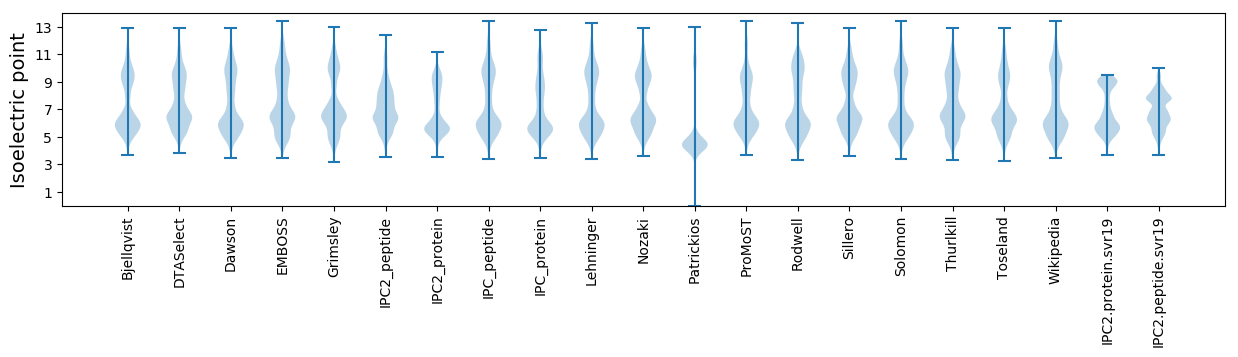
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B6RZU9|A0A0B6RZU9_BURPL Chaperone SurA OS=Burkholderia plantarii OX=41899 GN=surA PE=3 SV=1
MM1 pKa = 7.78SYY3 pKa = 7.45TTPSSLSDD11 pKa = 3.07FMGRR15 pKa = 11.84FEE17 pKa = 5.28NGGLLSGNTSGGTTTFYY34 pKa = 11.33YY35 pKa = 10.53EE36 pKa = 4.4PSTNTINFGYY46 pKa = 10.4GINLSFASGSTQARR60 pKa = 11.84GVIDD64 pKa = 3.81SALSAANISLSDD76 pKa = 3.96ALWSKK81 pKa = 10.51IKK83 pKa = 10.66NATAATRR90 pKa = 11.84GALVSDD96 pKa = 4.58LNNLVTNDD104 pKa = 3.47GSSPDD109 pKa = 3.56WQAVAASVTSAYY121 pKa = 7.59TQQIVVPEE129 pKa = 4.32LQSHH133 pKa = 6.36IPNLASLPATTQWALEE149 pKa = 4.02DD150 pKa = 3.61AFYY153 pKa = 11.19NSPSLLGPLVYY164 pKa = 10.31GAANSGNLASIAVQLAFNEE183 pKa = 4.33EE184 pKa = 4.29TPNAAQSGAEE194 pKa = 3.98LRR196 pKa = 11.84NLARR200 pKa = 11.84AALALGGSITFAPGGNTISSIDD222 pKa = 3.4WSKK225 pKa = 11.94ADD227 pKa = 3.55IASVGQFLSQIVSSAPVGKK246 pKa = 10.57GGSTADD252 pKa = 3.19AYY254 pKa = 9.67VNKK257 pKa = 9.2WGSSASASYY266 pKa = 8.76LTLTDD271 pKa = 3.2SLYY274 pKa = 11.04SYY276 pKa = 11.24VEE278 pKa = 3.8SQGRR282 pKa = 11.84YY283 pKa = 7.99VVDD286 pKa = 3.61TGQATYY292 pKa = 11.21ASIVSQNSDD301 pKa = 3.55DD302 pKa = 4.26PNFGNLTAADD312 pKa = 3.49IAALNGMSVSATLTAGQVIDD332 pKa = 4.03IPTSVTEE339 pKa = 4.27GLPASLSSSLSYY351 pKa = 10.53TLDD354 pKa = 3.2VTTGRR359 pKa = 11.84VVASPITDD367 pKa = 3.07GMFGGSVTVQNSATGAFLGYY387 pKa = 9.78FDD389 pKa = 5.51AGTVTGEE396 pKa = 3.54AAAASGQWTLSLNGGRR412 pKa = 11.84SVTFNADD419 pKa = 2.9SSISVSDD426 pKa = 3.8PAGSGSNFAPGASINVSSTGSATVLSSPGGTFSPVFVPTGNAYY469 pKa = 8.22GTDD472 pKa = 3.38LSTINSGQDD481 pKa = 2.7VGLATDD487 pKa = 3.96ISALMGIAAPIAIDD501 pKa = 4.87AVALDD506 pKa = 4.02YY507 pKa = 11.41LSDD510 pKa = 4.99RR511 pKa = 11.84IQYY514 pKa = 11.17SNVSDD519 pKa = 3.1KK520 pKa = 10.48WVAAFIGPAPEE531 pKa = 4.44GTFSLSSTLLSSTFDD546 pKa = 3.37PTTAALIEE554 pKa = 4.39LKK556 pKa = 9.46NTIANRR562 pKa = 11.84PKK564 pKa = 8.84PTWVSISIDD573 pKa = 3.29VTGFNSVPKK582 pKa = 9.23TSASVGILQQTDD594 pKa = 3.34AQVSPGTSLVQATMAGSQEE613 pKa = 3.83AAQVLNAYY621 pKa = 8.97INKK624 pKa = 9.68GYY626 pKa = 10.42AAAGAQTATSIATTLSGEE644 pKa = 4.3PTDD647 pKa = 4.12FTSPIAKK654 pKa = 8.76IQSWYY659 pKa = 8.3GTTRR663 pKa = 11.84IQGAGNGSTIGPTAGVPDD681 pKa = 5.24EE682 pKa = 3.98ISAYY686 pKa = 10.45GSTAVAGAVPSSNGGYY702 pKa = 9.76LIHH705 pKa = 6.4STAPADD711 pKa = 3.83TVRR714 pKa = 11.84ALQDD718 pKa = 3.44IQSLTPTAVAVARR731 pKa = 11.84SVAGGATAFGAAEE744 pKa = 4.42FAAVQADD751 pKa = 3.72SDD753 pKa = 3.94GSAASEE759 pKa = 3.67ATARR763 pKa = 11.84NLAVFANQAWGSALTEE779 pKa = 3.97YY780 pKa = 10.82LQVAEE785 pKa = 4.8NVSAMSQEE793 pKa = 3.96LSNVSAEE800 pKa = 4.17LTALEE805 pKa = 4.16PVSYY809 pKa = 10.13EE810 pKa = 3.74VTGNLVNGYY819 pKa = 9.67SYY821 pKa = 11.31YY822 pKa = 10.91SSGDD826 pKa = 3.19AAFAADD832 pKa = 4.27TFNGFADD839 pKa = 3.84GMKK842 pKa = 9.81TFAATKK848 pKa = 10.26DD849 pKa = 3.71GLDD852 pKa = 3.41RR853 pKa = 11.84ALLAIAQADD862 pKa = 3.59NASRR866 pKa = 11.84IFVGEE871 pKa = 4.15NGSSTTLEE879 pKa = 3.94SDD881 pKa = 2.8TDD883 pKa = 3.55IFVAGGGNEE892 pKa = 4.07TVADD896 pKa = 3.99GTGQDD901 pKa = 4.08SILVSSGSGNLYY913 pKa = 10.0ISNFKK918 pKa = 10.86AGINGDD924 pKa = 3.47QLQFLGVGDD933 pKa = 4.11NVVASDD939 pKa = 4.41DD940 pKa = 4.3GNGGTLLKK948 pKa = 10.6YY949 pKa = 10.81GSGNFIDD956 pKa = 5.64LIGVSAGSLNLYY968 pKa = 10.06QNITGVSAISFVGVSSPASFGLGGNRR994 pKa = 11.84IYY996 pKa = 10.87DD997 pKa = 3.9GQVHH1001 pKa = 6.55ISSITGTNSGSTLSGDD1017 pKa = 3.75SAATTLTGGAGNDD1030 pKa = 3.49TFKK1033 pKa = 11.41VEE1035 pKa = 4.02GNGYY1039 pKa = 9.35VINGAGGTNTVSYY1052 pKa = 10.82GDD1054 pKa = 3.56VPFGVSVNLQTGTDD1068 pKa = 3.58NLGTTLFAIQNVEE1081 pKa = 3.69GSSYY1085 pKa = 11.44DD1086 pKa = 3.45DD1087 pKa = 4.39TIAGDD1092 pKa = 3.73TNNNVLDD1099 pKa = 4.32GKK1101 pKa = 10.71GGNDD1105 pKa = 3.44TLIGGGGNDD1114 pKa = 3.52TYY1116 pKa = 11.68VFDD1119 pKa = 4.73AGYY1122 pKa = 11.0GSVTIEE1128 pKa = 4.75NGLAPNGIPSSEE1140 pKa = 4.14LLFGAGIDD1148 pKa = 5.37ANDD1151 pKa = 3.1LWFTRR1156 pKa = 11.84SGNDD1160 pKa = 2.98LTIQVLGTDD1169 pKa = 3.28NQVTVKK1175 pKa = 10.25GWFSNAWQQLASISLSDD1192 pKa = 3.88GLQVSPQTVATILKK1206 pKa = 9.51LQALYY1211 pKa = 10.72VAANPGFDD1219 pKa = 4.49PDD1221 pKa = 3.94TAQAMPGGIDD1231 pKa = 3.23LTSYY1235 pKa = 10.21FGPVSNPTTVPTATNVALATEE1256 pKa = 4.28HH1257 pKa = 7.03LYY1259 pKa = 10.61TSGVATQGANLAGSAATQINNPVGSAWSANVYY1291 pKa = 10.75ANNLAYY1297 pKa = 10.08GSKK1300 pKa = 10.48ALPAPYY1306 pKa = 10.37GSGEE1310 pKa = 4.04NYY1312 pKa = 10.02AYY1314 pKa = 10.22QYY1316 pKa = 10.94YY1317 pKa = 8.24VTGSSQPYY1325 pKa = 7.73VALSVRR1331 pKa = 11.84PWGQYY1336 pKa = 10.96APGTSISTTPPGVTSYY1352 pKa = 10.07QQIYY1356 pKa = 10.45ALTPSTFYY1364 pKa = 11.41VKK1366 pKa = 10.04ATTSVSPSSSSNPDD1380 pKa = 2.84SSLVFASDD1388 pKa = 3.59PNVMVNAINAVTEE1401 pKa = 4.17AVDD1404 pKa = 4.03LATTGATLAQAISTADD1420 pKa = 3.55TARR1423 pKa = 11.84QTALGSAVAANSAGAAFGSQPAVQARR1449 pKa = 11.84ADD1451 pKa = 3.58ATTAEE1456 pKa = 4.44ADD1458 pKa = 3.58LAAALAQWAPVQSNLVTAGGVLASSQAALNGILPGTQVSTQQRR1501 pKa = 11.84TGYY1504 pKa = 9.63GLSGPSTVTVTYY1516 pKa = 6.11TTNYY1520 pKa = 10.25SFYY1523 pKa = 10.18TSTDD1527 pKa = 3.1QAEE1530 pKa = 4.29FNALQSAVSNAQGAVGEE1547 pKa = 4.62ADD1549 pKa = 3.83FLMLSTLQSALQDD1562 pKa = 3.63FGAYY1566 pKa = 9.49EE1567 pKa = 4.09RR1568 pKa = 11.84VQVGGTSSNLGADD1581 pKa = 3.73DD1582 pKa = 5.68GGDD1585 pKa = 3.71LLIAAGGGYY1594 pKa = 8.09HH1595 pKa = 5.48TVVGGAGRR1603 pKa = 11.84DD1604 pKa = 3.46TFVFGGWNDD1613 pKa = 3.44TSSATVQNFATGTQGDD1629 pKa = 4.32RR1630 pKa = 11.84LLLIPAQNRR1639 pKa = 11.84TVYY1642 pKa = 9.44LTDD1645 pKa = 3.78GGGTTDD1651 pKa = 4.29ASFFVDD1657 pKa = 3.2NGGVDD1662 pKa = 3.91TVALAGVSLGALSLYY1677 pKa = 11.14DD1678 pKa = 3.63NFNGVDD1684 pKa = 3.23TASFANEE1691 pKa = 3.82SHH1693 pKa = 6.19GVSIDD1698 pKa = 3.47LATVTPRR1705 pKa = 11.84SYY1707 pKa = 11.23DD1708 pKa = 3.03GSTHH1712 pKa = 5.3IRR1714 pKa = 11.84NLVGSNYY1721 pKa = 10.32GDD1723 pKa = 3.62TLTGDD1728 pKa = 3.58EE1729 pKa = 4.42QDD1731 pKa = 3.56NVITGGAGNDD1741 pKa = 3.73VLSGGAGNNTLDD1753 pKa = 4.25GGGGNNTVSYY1763 pKa = 9.86AGSPGGVTVDD1773 pKa = 3.86LTAGTASNGYY1783 pKa = 10.0GGTDD1787 pKa = 3.32TLSHH1791 pKa = 5.92IQNVIGSGGNDD1802 pKa = 3.19TMFGDD1807 pKa = 4.28ANNNVLDD1814 pKa = 4.34GNGGNDD1820 pKa = 3.32VLVGGGGSDD1829 pKa = 3.31TYY1831 pKa = 11.63VFDD1834 pKa = 4.48AGYY1837 pKa = 9.48GQDD1840 pKa = 3.54VIVNGVAGNAGPSGQLTLGAGLSAANLRR1868 pKa = 11.84FVQSGNDD1875 pKa = 3.41LVIEE1879 pKa = 4.13VLGTTEE1885 pKa = 4.03QVTVQDD1891 pKa = 3.84WFQAGASYY1899 pKa = 10.56RR1900 pKa = 11.84QLATIALADD1909 pKa = 3.84GTTLSMTDD1917 pKa = 3.21VNGLVAEE1924 pKa = 4.35GHH1926 pKa = 6.69DD1927 pKa = 4.18WTPLSTPSASTSPTLAGRR1945 pKa = 11.84YY1946 pKa = 8.82VYY1948 pKa = 10.02IRR1950 pKa = 11.84KK1951 pKa = 9.3NDD1953 pKa = 3.65GASTGLVTSEE1963 pKa = 3.64IQVWGTDD1970 pKa = 3.5GSDD1973 pKa = 3.22LALGKK1978 pKa = 10.29AVTSSPSWGDD1988 pKa = 2.85GTTAVNAVNGSTSASGGIFASASADD2013 pKa = 2.79GWVEE2017 pKa = 3.59VDD2019 pKa = 3.89LGSVQTIAGVDD2030 pKa = 2.94VWGRR2034 pKa = 11.84DD2035 pKa = 3.62DD2036 pKa = 5.56GSEE2039 pKa = 4.05EE2040 pKa = 3.9MNGNYY2045 pKa = 8.4TVYY2048 pKa = 10.74VSNQDD2053 pKa = 3.16MGGLDD2058 pKa = 3.73AGQLQAVPGARR2069 pKa = 11.84AYY2071 pKa = 10.35QEE2073 pKa = 4.16NQPGPMDD2080 pKa = 3.56VKK2082 pKa = 9.52VTSWQAPSTVTQTTGRR2098 pKa = 11.84YY2099 pKa = 8.82VYY2101 pKa = 10.43VRR2103 pKa = 11.84KK2104 pKa = 10.0NDD2106 pKa = 3.65GASTGLVTSEE2116 pKa = 3.74IQVWGAGGVDD2126 pKa = 3.66LALGKK2131 pKa = 8.44TVTSSSSWSSDD2142 pKa = 2.62TTAANAINGVTSASGGLFASASLDD2166 pKa = 3.03GWIEE2170 pKa = 3.67IDD2172 pKa = 3.81LGSVQAIDD2180 pKa = 5.94GIDD2183 pKa = 2.9VWGRR2187 pKa = 11.84DD2188 pKa = 3.36DD2189 pKa = 5.73GAFEE2193 pKa = 4.27MNGDD2197 pKa = 3.53YY2198 pKa = 10.24TVYY2201 pKa = 10.79VSNQDD2206 pKa = 3.73LAGLDD2211 pKa = 3.4ASQIQASVPNVQRR2224 pKa = 11.84FQQNDD2229 pKa = 3.34RR2230 pKa = 11.84GPNDD2234 pKa = 3.38VAVSSWQAPTALAATAGRR2252 pKa = 11.84YY2253 pKa = 9.12VYY2255 pKa = 10.45VRR2257 pKa = 11.84KK2258 pKa = 10.0NDD2260 pKa = 3.65GASTGLVTSEE2270 pKa = 3.8IQVWGADD2277 pKa = 3.61GSDD2280 pKa = 3.23LALGKK2285 pKa = 8.41TVTSSTSWGDD2295 pKa = 3.15STMAANAVNGSTGASGGIFASASADD2320 pKa = 2.83GWIEE2324 pKa = 3.73VDD2326 pKa = 3.64LGSVQTINGIDD2337 pKa = 3.0VWGRR2341 pKa = 11.84DD2342 pKa = 4.31DD2343 pKa = 5.46GALDD2347 pKa = 3.39MNGNYY2352 pKa = 8.38TVYY2355 pKa = 10.8VSNQDD2360 pKa = 3.75LAGLSAAQAQATAGVKK2376 pKa = 9.66QFQQSQQSQQGALDD2390 pKa = 3.71TAVTSWQTPATAATTLGRR2408 pKa = 11.84YY2409 pKa = 9.26VYY2411 pKa = 10.43VRR2413 pKa = 11.84KK2414 pKa = 10.0NDD2416 pKa = 3.65GASTGLVTSEE2426 pKa = 3.64IQVWGTDD2433 pKa = 3.5GSDD2436 pKa = 3.11LALGKK2441 pKa = 8.45TVTSSKK2447 pKa = 10.5SWGDD2451 pKa = 3.19STTAANAINGVTSASGGIFASASADD2476 pKa = 2.82GWVEE2480 pKa = 3.42IDD2482 pKa = 4.41LGSAQTIAGIDD2493 pKa = 3.04VWGRR2497 pKa = 11.84DD2498 pKa = 4.31DD2499 pKa = 5.46GALDD2503 pKa = 3.39MNGNYY2508 pKa = 8.89TVFVSNQDD2516 pKa = 3.46MAGLDD2521 pKa = 3.56ANQIQSISGAKK2532 pKa = 9.94AFQQNQQGPLDD2543 pKa = 3.75VSVTSWQSAAGSMVQAMAAFSSSSAGATNTTFVPQTQQSSMLAASMQQ2590 pKa = 3.38
MM1 pKa = 7.78SYY3 pKa = 7.45TTPSSLSDD11 pKa = 3.07FMGRR15 pKa = 11.84FEE17 pKa = 5.28NGGLLSGNTSGGTTTFYY34 pKa = 11.33YY35 pKa = 10.53EE36 pKa = 4.4PSTNTINFGYY46 pKa = 10.4GINLSFASGSTQARR60 pKa = 11.84GVIDD64 pKa = 3.81SALSAANISLSDD76 pKa = 3.96ALWSKK81 pKa = 10.51IKK83 pKa = 10.66NATAATRR90 pKa = 11.84GALVSDD96 pKa = 4.58LNNLVTNDD104 pKa = 3.47GSSPDD109 pKa = 3.56WQAVAASVTSAYY121 pKa = 7.59TQQIVVPEE129 pKa = 4.32LQSHH133 pKa = 6.36IPNLASLPATTQWALEE149 pKa = 4.02DD150 pKa = 3.61AFYY153 pKa = 11.19NSPSLLGPLVYY164 pKa = 10.31GAANSGNLASIAVQLAFNEE183 pKa = 4.33EE184 pKa = 4.29TPNAAQSGAEE194 pKa = 3.98LRR196 pKa = 11.84NLARR200 pKa = 11.84AALALGGSITFAPGGNTISSIDD222 pKa = 3.4WSKK225 pKa = 11.94ADD227 pKa = 3.55IASVGQFLSQIVSSAPVGKK246 pKa = 10.57GGSTADD252 pKa = 3.19AYY254 pKa = 9.67VNKK257 pKa = 9.2WGSSASASYY266 pKa = 8.76LTLTDD271 pKa = 3.2SLYY274 pKa = 11.04SYY276 pKa = 11.24VEE278 pKa = 3.8SQGRR282 pKa = 11.84YY283 pKa = 7.99VVDD286 pKa = 3.61TGQATYY292 pKa = 11.21ASIVSQNSDD301 pKa = 3.55DD302 pKa = 4.26PNFGNLTAADD312 pKa = 3.49IAALNGMSVSATLTAGQVIDD332 pKa = 4.03IPTSVTEE339 pKa = 4.27GLPASLSSSLSYY351 pKa = 10.53TLDD354 pKa = 3.2VTTGRR359 pKa = 11.84VVASPITDD367 pKa = 3.07GMFGGSVTVQNSATGAFLGYY387 pKa = 9.78FDD389 pKa = 5.51AGTVTGEE396 pKa = 3.54AAAASGQWTLSLNGGRR412 pKa = 11.84SVTFNADD419 pKa = 2.9SSISVSDD426 pKa = 3.8PAGSGSNFAPGASINVSSTGSATVLSSPGGTFSPVFVPTGNAYY469 pKa = 8.22GTDD472 pKa = 3.38LSTINSGQDD481 pKa = 2.7VGLATDD487 pKa = 3.96ISALMGIAAPIAIDD501 pKa = 4.87AVALDD506 pKa = 4.02YY507 pKa = 11.41LSDD510 pKa = 4.99RR511 pKa = 11.84IQYY514 pKa = 11.17SNVSDD519 pKa = 3.1KK520 pKa = 10.48WVAAFIGPAPEE531 pKa = 4.44GTFSLSSTLLSSTFDD546 pKa = 3.37PTTAALIEE554 pKa = 4.39LKK556 pKa = 9.46NTIANRR562 pKa = 11.84PKK564 pKa = 8.84PTWVSISIDD573 pKa = 3.29VTGFNSVPKK582 pKa = 9.23TSASVGILQQTDD594 pKa = 3.34AQVSPGTSLVQATMAGSQEE613 pKa = 3.83AAQVLNAYY621 pKa = 8.97INKK624 pKa = 9.68GYY626 pKa = 10.42AAAGAQTATSIATTLSGEE644 pKa = 4.3PTDD647 pKa = 4.12FTSPIAKK654 pKa = 8.76IQSWYY659 pKa = 8.3GTTRR663 pKa = 11.84IQGAGNGSTIGPTAGVPDD681 pKa = 5.24EE682 pKa = 3.98ISAYY686 pKa = 10.45GSTAVAGAVPSSNGGYY702 pKa = 9.76LIHH705 pKa = 6.4STAPADD711 pKa = 3.83TVRR714 pKa = 11.84ALQDD718 pKa = 3.44IQSLTPTAVAVARR731 pKa = 11.84SVAGGATAFGAAEE744 pKa = 4.42FAAVQADD751 pKa = 3.72SDD753 pKa = 3.94GSAASEE759 pKa = 3.67ATARR763 pKa = 11.84NLAVFANQAWGSALTEE779 pKa = 3.97YY780 pKa = 10.82LQVAEE785 pKa = 4.8NVSAMSQEE793 pKa = 3.96LSNVSAEE800 pKa = 4.17LTALEE805 pKa = 4.16PVSYY809 pKa = 10.13EE810 pKa = 3.74VTGNLVNGYY819 pKa = 9.67SYY821 pKa = 11.31YY822 pKa = 10.91SSGDD826 pKa = 3.19AAFAADD832 pKa = 4.27TFNGFADD839 pKa = 3.84GMKK842 pKa = 9.81TFAATKK848 pKa = 10.26DD849 pKa = 3.71GLDD852 pKa = 3.41RR853 pKa = 11.84ALLAIAQADD862 pKa = 3.59NASRR866 pKa = 11.84IFVGEE871 pKa = 4.15NGSSTTLEE879 pKa = 3.94SDD881 pKa = 2.8TDD883 pKa = 3.55IFVAGGGNEE892 pKa = 4.07TVADD896 pKa = 3.99GTGQDD901 pKa = 4.08SILVSSGSGNLYY913 pKa = 10.0ISNFKK918 pKa = 10.86AGINGDD924 pKa = 3.47QLQFLGVGDD933 pKa = 4.11NVVASDD939 pKa = 4.41DD940 pKa = 4.3GNGGTLLKK948 pKa = 10.6YY949 pKa = 10.81GSGNFIDD956 pKa = 5.64LIGVSAGSLNLYY968 pKa = 10.06QNITGVSAISFVGVSSPASFGLGGNRR994 pKa = 11.84IYY996 pKa = 10.87DD997 pKa = 3.9GQVHH1001 pKa = 6.55ISSITGTNSGSTLSGDD1017 pKa = 3.75SAATTLTGGAGNDD1030 pKa = 3.49TFKK1033 pKa = 11.41VEE1035 pKa = 4.02GNGYY1039 pKa = 9.35VINGAGGTNTVSYY1052 pKa = 10.82GDD1054 pKa = 3.56VPFGVSVNLQTGTDD1068 pKa = 3.58NLGTTLFAIQNVEE1081 pKa = 3.69GSSYY1085 pKa = 11.44DD1086 pKa = 3.45DD1087 pKa = 4.39TIAGDD1092 pKa = 3.73TNNNVLDD1099 pKa = 4.32GKK1101 pKa = 10.71GGNDD1105 pKa = 3.44TLIGGGGNDD1114 pKa = 3.52TYY1116 pKa = 11.68VFDD1119 pKa = 4.73AGYY1122 pKa = 11.0GSVTIEE1128 pKa = 4.75NGLAPNGIPSSEE1140 pKa = 4.14LLFGAGIDD1148 pKa = 5.37ANDD1151 pKa = 3.1LWFTRR1156 pKa = 11.84SGNDD1160 pKa = 2.98LTIQVLGTDD1169 pKa = 3.28NQVTVKK1175 pKa = 10.25GWFSNAWQQLASISLSDD1192 pKa = 3.88GLQVSPQTVATILKK1206 pKa = 9.51LQALYY1211 pKa = 10.72VAANPGFDD1219 pKa = 4.49PDD1221 pKa = 3.94TAQAMPGGIDD1231 pKa = 3.23LTSYY1235 pKa = 10.21FGPVSNPTTVPTATNVALATEE1256 pKa = 4.28HH1257 pKa = 7.03LYY1259 pKa = 10.61TSGVATQGANLAGSAATQINNPVGSAWSANVYY1291 pKa = 10.75ANNLAYY1297 pKa = 10.08GSKK1300 pKa = 10.48ALPAPYY1306 pKa = 10.37GSGEE1310 pKa = 4.04NYY1312 pKa = 10.02AYY1314 pKa = 10.22QYY1316 pKa = 10.94YY1317 pKa = 8.24VTGSSQPYY1325 pKa = 7.73VALSVRR1331 pKa = 11.84PWGQYY1336 pKa = 10.96APGTSISTTPPGVTSYY1352 pKa = 10.07QQIYY1356 pKa = 10.45ALTPSTFYY1364 pKa = 11.41VKK1366 pKa = 10.04ATTSVSPSSSSNPDD1380 pKa = 2.84SSLVFASDD1388 pKa = 3.59PNVMVNAINAVTEE1401 pKa = 4.17AVDD1404 pKa = 4.03LATTGATLAQAISTADD1420 pKa = 3.55TARR1423 pKa = 11.84QTALGSAVAANSAGAAFGSQPAVQARR1449 pKa = 11.84ADD1451 pKa = 3.58ATTAEE1456 pKa = 4.44ADD1458 pKa = 3.58LAAALAQWAPVQSNLVTAGGVLASSQAALNGILPGTQVSTQQRR1501 pKa = 11.84TGYY1504 pKa = 9.63GLSGPSTVTVTYY1516 pKa = 6.11TTNYY1520 pKa = 10.25SFYY1523 pKa = 10.18TSTDD1527 pKa = 3.1QAEE1530 pKa = 4.29FNALQSAVSNAQGAVGEE1547 pKa = 4.62ADD1549 pKa = 3.83FLMLSTLQSALQDD1562 pKa = 3.63FGAYY1566 pKa = 9.49EE1567 pKa = 4.09RR1568 pKa = 11.84VQVGGTSSNLGADD1581 pKa = 3.73DD1582 pKa = 5.68GGDD1585 pKa = 3.71LLIAAGGGYY1594 pKa = 8.09HH1595 pKa = 5.48TVVGGAGRR1603 pKa = 11.84DD1604 pKa = 3.46TFVFGGWNDD1613 pKa = 3.44TSSATVQNFATGTQGDD1629 pKa = 4.32RR1630 pKa = 11.84LLLIPAQNRR1639 pKa = 11.84TVYY1642 pKa = 9.44LTDD1645 pKa = 3.78GGGTTDD1651 pKa = 4.29ASFFVDD1657 pKa = 3.2NGGVDD1662 pKa = 3.91TVALAGVSLGALSLYY1677 pKa = 11.14DD1678 pKa = 3.63NFNGVDD1684 pKa = 3.23TASFANEE1691 pKa = 3.82SHH1693 pKa = 6.19GVSIDD1698 pKa = 3.47LATVTPRR1705 pKa = 11.84SYY1707 pKa = 11.23DD1708 pKa = 3.03GSTHH1712 pKa = 5.3IRR1714 pKa = 11.84NLVGSNYY1721 pKa = 10.32GDD1723 pKa = 3.62TLTGDD1728 pKa = 3.58EE1729 pKa = 4.42QDD1731 pKa = 3.56NVITGGAGNDD1741 pKa = 3.73VLSGGAGNNTLDD1753 pKa = 4.25GGGGNNTVSYY1763 pKa = 9.86AGSPGGVTVDD1773 pKa = 3.86LTAGTASNGYY1783 pKa = 10.0GGTDD1787 pKa = 3.32TLSHH1791 pKa = 5.92IQNVIGSGGNDD1802 pKa = 3.19TMFGDD1807 pKa = 4.28ANNNVLDD1814 pKa = 4.34GNGGNDD1820 pKa = 3.32VLVGGGGSDD1829 pKa = 3.31TYY1831 pKa = 11.63VFDD1834 pKa = 4.48AGYY1837 pKa = 9.48GQDD1840 pKa = 3.54VIVNGVAGNAGPSGQLTLGAGLSAANLRR1868 pKa = 11.84FVQSGNDD1875 pKa = 3.41LVIEE1879 pKa = 4.13VLGTTEE1885 pKa = 4.03QVTVQDD1891 pKa = 3.84WFQAGASYY1899 pKa = 10.56RR1900 pKa = 11.84QLATIALADD1909 pKa = 3.84GTTLSMTDD1917 pKa = 3.21VNGLVAEE1924 pKa = 4.35GHH1926 pKa = 6.69DD1927 pKa = 4.18WTPLSTPSASTSPTLAGRR1945 pKa = 11.84YY1946 pKa = 8.82VYY1948 pKa = 10.02IRR1950 pKa = 11.84KK1951 pKa = 9.3NDD1953 pKa = 3.65GASTGLVTSEE1963 pKa = 3.64IQVWGTDD1970 pKa = 3.5GSDD1973 pKa = 3.22LALGKK1978 pKa = 10.29AVTSSPSWGDD1988 pKa = 2.85GTTAVNAVNGSTSASGGIFASASADD2013 pKa = 2.79GWVEE2017 pKa = 3.59VDD2019 pKa = 3.89LGSVQTIAGVDD2030 pKa = 2.94VWGRR2034 pKa = 11.84DD2035 pKa = 3.62DD2036 pKa = 5.56GSEE2039 pKa = 4.05EE2040 pKa = 3.9MNGNYY2045 pKa = 8.4TVYY2048 pKa = 10.74VSNQDD2053 pKa = 3.16MGGLDD2058 pKa = 3.73AGQLQAVPGARR2069 pKa = 11.84AYY2071 pKa = 10.35QEE2073 pKa = 4.16NQPGPMDD2080 pKa = 3.56VKK2082 pKa = 9.52VTSWQAPSTVTQTTGRR2098 pKa = 11.84YY2099 pKa = 8.82VYY2101 pKa = 10.43VRR2103 pKa = 11.84KK2104 pKa = 10.0NDD2106 pKa = 3.65GASTGLVTSEE2116 pKa = 3.74IQVWGAGGVDD2126 pKa = 3.66LALGKK2131 pKa = 8.44TVTSSSSWSSDD2142 pKa = 2.62TTAANAINGVTSASGGLFASASLDD2166 pKa = 3.03GWIEE2170 pKa = 3.67IDD2172 pKa = 3.81LGSVQAIDD2180 pKa = 5.94GIDD2183 pKa = 2.9VWGRR2187 pKa = 11.84DD2188 pKa = 3.36DD2189 pKa = 5.73GAFEE2193 pKa = 4.27MNGDD2197 pKa = 3.53YY2198 pKa = 10.24TVYY2201 pKa = 10.79VSNQDD2206 pKa = 3.73LAGLDD2211 pKa = 3.4ASQIQASVPNVQRR2224 pKa = 11.84FQQNDD2229 pKa = 3.34RR2230 pKa = 11.84GPNDD2234 pKa = 3.38VAVSSWQAPTALAATAGRR2252 pKa = 11.84YY2253 pKa = 9.12VYY2255 pKa = 10.45VRR2257 pKa = 11.84KK2258 pKa = 10.0NDD2260 pKa = 3.65GASTGLVTSEE2270 pKa = 3.8IQVWGADD2277 pKa = 3.61GSDD2280 pKa = 3.23LALGKK2285 pKa = 8.41TVTSSTSWGDD2295 pKa = 3.15STMAANAVNGSTGASGGIFASASADD2320 pKa = 2.83GWIEE2324 pKa = 3.73VDD2326 pKa = 3.64LGSVQTINGIDD2337 pKa = 3.0VWGRR2341 pKa = 11.84DD2342 pKa = 4.31DD2343 pKa = 5.46GALDD2347 pKa = 3.39MNGNYY2352 pKa = 8.38TVYY2355 pKa = 10.8VSNQDD2360 pKa = 3.75LAGLSAAQAQATAGVKK2376 pKa = 9.66QFQQSQQSQQGALDD2390 pKa = 3.71TAVTSWQTPATAATTLGRR2408 pKa = 11.84YY2409 pKa = 9.26VYY2411 pKa = 10.43VRR2413 pKa = 11.84KK2414 pKa = 10.0NDD2416 pKa = 3.65GASTGLVTSEE2426 pKa = 3.64IQVWGTDD2433 pKa = 3.5GSDD2436 pKa = 3.11LALGKK2441 pKa = 8.45TVTSSKK2447 pKa = 10.5SWGDD2451 pKa = 3.19STTAANAINGVTSASGGIFASASADD2476 pKa = 2.82GWVEE2480 pKa = 3.42IDD2482 pKa = 4.41LGSAQTIAGIDD2493 pKa = 3.04VWGRR2497 pKa = 11.84DD2498 pKa = 4.31DD2499 pKa = 5.46GALDD2503 pKa = 3.39MNGNYY2508 pKa = 8.89TVFVSNQDD2516 pKa = 3.46MAGLDD2521 pKa = 3.56ANQIQSISGAKK2532 pKa = 9.94AFQQNQQGPLDD2543 pKa = 3.75VSVTSWQSAAGSMVQAMAAFSSSSAGATNTTFVPQTQQSSMLAASMQQ2590 pKa = 3.38
Molecular weight: 261.92 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B6RS76|A0A0B6RS76_BURPL Uncharacterized protein OS=Burkholderia plantarii OX=41899 GN=BGL_2c01060 PE=4 SV=1
MM1 pKa = 7.59SFRR4 pKa = 11.84LTSRR8 pKa = 11.84LHH10 pKa = 5.28RR11 pKa = 11.84AARR14 pKa = 11.84RR15 pKa = 11.84LSQLEE20 pKa = 3.55PRR22 pKa = 11.84RR23 pKa = 11.84IVRR26 pKa = 11.84TLRR29 pKa = 11.84HH30 pKa = 4.88HH31 pKa = 5.53QARR34 pKa = 11.84TRR36 pKa = 11.84ALATRR41 pKa = 11.84AVASTPRR48 pKa = 11.84LDD50 pKa = 4.77GLRR53 pKa = 11.84AWLHH57 pKa = 5.75SFLAAMRR64 pKa = 11.84AHH66 pKa = 6.93AGARR70 pKa = 11.84RR71 pKa = 11.84LGLPALLRR79 pKa = 11.84KK80 pKa = 9.51PSSLRR85 pKa = 11.84LLALRR90 pKa = 11.84RR91 pKa = 11.84ANGPRR96 pKa = 11.84RR97 pKa = 11.84RR98 pKa = 11.84VNRR101 pKa = 11.84PRR103 pKa = 11.84RR104 pKa = 11.84LSASPGWFAFGARR117 pKa = 3.97
MM1 pKa = 7.59SFRR4 pKa = 11.84LTSRR8 pKa = 11.84LHH10 pKa = 5.28RR11 pKa = 11.84AARR14 pKa = 11.84RR15 pKa = 11.84LSQLEE20 pKa = 3.55PRR22 pKa = 11.84RR23 pKa = 11.84IVRR26 pKa = 11.84TLRR29 pKa = 11.84HH30 pKa = 4.88HH31 pKa = 5.53QARR34 pKa = 11.84TRR36 pKa = 11.84ALATRR41 pKa = 11.84AVASTPRR48 pKa = 11.84LDD50 pKa = 4.77GLRR53 pKa = 11.84AWLHH57 pKa = 5.75SFLAAMRR64 pKa = 11.84AHH66 pKa = 6.93AGARR70 pKa = 11.84RR71 pKa = 11.84LGLPALLRR79 pKa = 11.84KK80 pKa = 9.51PSSLRR85 pKa = 11.84LLALRR90 pKa = 11.84RR91 pKa = 11.84ANGPRR96 pKa = 11.84RR97 pKa = 11.84RR98 pKa = 11.84VNRR101 pKa = 11.84PRR103 pKa = 11.84RR104 pKa = 11.84LSASPGWFAFGARR117 pKa = 3.97
Molecular weight: 13.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2225259 |
44 |
7858 |
346.0 |
37.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.097 ± 0.058 | 0.89 ± 0.011 |
5.595 ± 0.027 | 4.877 ± 0.035 |
3.529 ± 0.019 | 8.575 ± 0.039 |
2.341 ± 0.016 | 4.305 ± 0.024 |
2.426 ± 0.029 | 10.345 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.129 ± 0.016 | 2.472 ± 0.024 |
5.472 ± 0.029 | 3.321 ± 0.02 |
7.718 ± 0.044 | 5.393 ± 0.059 |
5.308 ± 0.037 | 7.516 ± 0.024 |
1.36 ± 0.012 | 2.333 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
