
Leptonychotes weddellii papillomavirus 5
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; unclassified Papillomaviridae; non-primate mammal papillomaviruses
Average proteome isoelectric point is 5.84
Get precalculated fractions of proteins
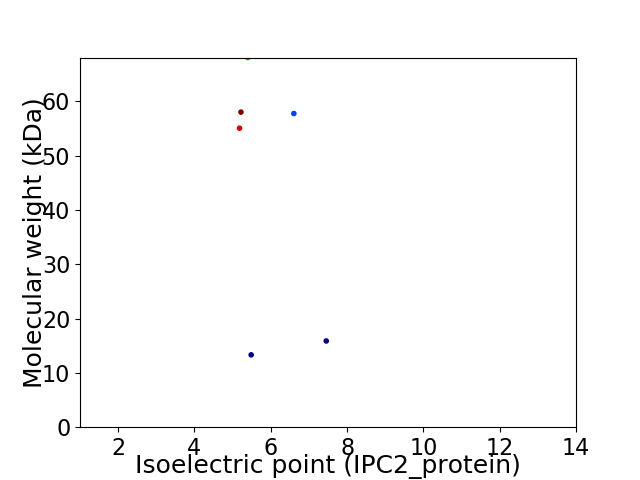
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2I8B2T9|A0A2I8B2T9_9PAPI Replication protein E1 OS=Leptonychotes weddellii papillomavirus 5 OX=2077306 GN=E1 PE=3 SV=1
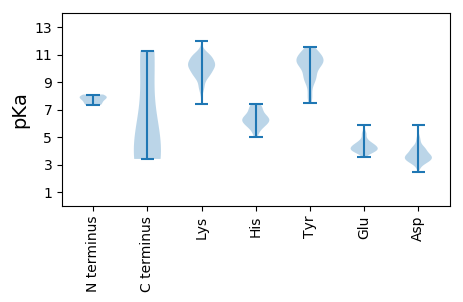
MM1 pKa = 7.75APRR4 pKa = 11.84AARR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 9.63RR10 pKa = 11.84ADD12 pKa = 3.1AGSLYY17 pKa = 9.95RR18 pKa = 11.84HH19 pKa = 6.46CVQGGDD25 pKa = 4.55CIPDD29 pKa = 3.28VVNKK33 pKa = 10.18FEE35 pKa = 5.09HH36 pKa = 6.18KK37 pKa = 9.55TPADD41 pKa = 4.87RR42 pKa = 11.84ILQIGGSLVYY52 pKa = 10.22FGGAGISTGRR62 pKa = 11.84GAGGYY67 pKa = 9.33RR68 pKa = 11.84PLGGTRR74 pKa = 11.84TFTFGSRR81 pKa = 11.84GAAGPGRR88 pKa = 11.84LPPLLADD95 pKa = 3.41TLGPVDD101 pKa = 3.36ATVYY105 pKa = 10.64EE106 pKa = 5.24GITFDD111 pKa = 3.8SPAVSVGVDD120 pKa = 3.32EE121 pKa = 5.44GPAIEE126 pKa = 4.48LTPIGGRR133 pKa = 11.84DD134 pKa = 3.72GQPGSGAPTISTTTDD149 pKa = 2.48SDD151 pKa = 4.06VAILEE156 pKa = 4.58VGTSTPSSRR165 pKa = 11.84PTSRR169 pKa = 11.84GGNVVSRR176 pKa = 11.84SQFEE180 pKa = 4.18NPTFVPTQPGTPGVGEE196 pKa = 4.74FSAQEE201 pKa = 4.18NILVDD206 pKa = 3.62HH207 pKa = 6.86SGGGVSVGEE216 pKa = 4.14FEE218 pKa = 4.93EE219 pKa = 5.75IEE221 pKa = 4.32LGDD224 pKa = 3.77LGEE227 pKa = 4.39RR228 pKa = 11.84TDD230 pKa = 3.53TGVPKK235 pKa = 10.49TSTPKK240 pKa = 10.42SRR242 pKa = 11.84LGDD245 pKa = 3.59LVSKK249 pKa = 10.89AKK251 pKa = 10.23SLYY254 pKa = 9.23NRR256 pKa = 11.84RR257 pKa = 11.84VTQTRR262 pKa = 11.84VEE264 pKa = 4.07NPEE267 pKa = 3.67FLGRR271 pKa = 11.84PSSLVEE277 pKa = 4.24FGFEE281 pKa = 3.87NPAFEE286 pKa = 5.31GDD288 pKa = 3.43VTLTFDD294 pKa = 4.18RR295 pKa = 11.84VQEE298 pKa = 4.06AKK300 pKa = 10.36AAPDD304 pKa = 3.3LAFRR308 pKa = 11.84DD309 pKa = 4.03IIKK312 pKa = 10.36LSRR315 pKa = 11.84PIYY318 pKa = 10.35SEE320 pKa = 4.21APGGRR325 pKa = 11.84VRR327 pKa = 11.84VSRR330 pKa = 11.84LGTKK334 pKa = 8.53GTIRR338 pKa = 11.84LRR340 pKa = 11.84SGTLIGGQTHH350 pKa = 6.1YY351 pKa = 10.82FRR353 pKa = 11.84DD354 pKa = 3.84LSSIGAPSLEE364 pKa = 4.03MSVLGQTSGEE374 pKa = 4.31STVVLGSDD382 pKa = 3.28SSIINGVGLEE392 pKa = 4.16GTVAFPEE399 pKa = 4.22EE400 pKa = 3.95QLLLDD405 pKa = 4.01EE406 pKa = 5.47LSEE409 pKa = 4.23NFQNSQLVFSDD420 pKa = 3.16SHH422 pKa = 6.4QSHH425 pKa = 7.19IIEE428 pKa = 4.55LPRR431 pKa = 11.84IAEE434 pKa = 4.41SPRR437 pKa = 11.84SSYY440 pKa = 11.04VIYY443 pKa = 8.8DD444 pKa = 3.8TADD447 pKa = 3.37STVVSHH453 pKa = 7.18KK454 pKa = 10.86GNEE457 pKa = 3.95NYY459 pKa = 10.28FISTLTPPDD468 pKa = 4.05TEE470 pKa = 5.23PDD472 pKa = 3.91TEE474 pKa = 4.26PQTPDD479 pKa = 3.01EE480 pKa = 4.47TPRR483 pKa = 11.84APRR486 pKa = 11.84RR487 pKa = 11.84PLDD490 pKa = 4.3DD491 pKa = 4.25SDD493 pKa = 5.1GLTFDD498 pKa = 4.33LHH500 pKa = 6.56PGLLRR505 pKa = 11.84RR506 pKa = 11.84KK507 pKa = 8.63KK508 pKa = 10.4KK509 pKa = 9.51RR510 pKa = 11.84VRR512 pKa = 11.84YY513 pKa = 9.54YY514 pKa = 10.22IEE516 pKa = 3.8
MM1 pKa = 7.75APRR4 pKa = 11.84AARR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 9.63RR10 pKa = 11.84ADD12 pKa = 3.1AGSLYY17 pKa = 9.95RR18 pKa = 11.84HH19 pKa = 6.46CVQGGDD25 pKa = 4.55CIPDD29 pKa = 3.28VVNKK33 pKa = 10.18FEE35 pKa = 5.09HH36 pKa = 6.18KK37 pKa = 9.55TPADD41 pKa = 4.87RR42 pKa = 11.84ILQIGGSLVYY52 pKa = 10.22FGGAGISTGRR62 pKa = 11.84GAGGYY67 pKa = 9.33RR68 pKa = 11.84PLGGTRR74 pKa = 11.84TFTFGSRR81 pKa = 11.84GAAGPGRR88 pKa = 11.84LPPLLADD95 pKa = 3.41TLGPVDD101 pKa = 3.36ATVYY105 pKa = 10.64EE106 pKa = 5.24GITFDD111 pKa = 3.8SPAVSVGVDD120 pKa = 3.32EE121 pKa = 5.44GPAIEE126 pKa = 4.48LTPIGGRR133 pKa = 11.84DD134 pKa = 3.72GQPGSGAPTISTTTDD149 pKa = 2.48SDD151 pKa = 4.06VAILEE156 pKa = 4.58VGTSTPSSRR165 pKa = 11.84PTSRR169 pKa = 11.84GGNVVSRR176 pKa = 11.84SQFEE180 pKa = 4.18NPTFVPTQPGTPGVGEE196 pKa = 4.74FSAQEE201 pKa = 4.18NILVDD206 pKa = 3.62HH207 pKa = 6.86SGGGVSVGEE216 pKa = 4.14FEE218 pKa = 4.93EE219 pKa = 5.75IEE221 pKa = 4.32LGDD224 pKa = 3.77LGEE227 pKa = 4.39RR228 pKa = 11.84TDD230 pKa = 3.53TGVPKK235 pKa = 10.49TSTPKK240 pKa = 10.42SRR242 pKa = 11.84LGDD245 pKa = 3.59LVSKK249 pKa = 10.89AKK251 pKa = 10.23SLYY254 pKa = 9.23NRR256 pKa = 11.84RR257 pKa = 11.84VTQTRR262 pKa = 11.84VEE264 pKa = 4.07NPEE267 pKa = 3.67FLGRR271 pKa = 11.84PSSLVEE277 pKa = 4.24FGFEE281 pKa = 3.87NPAFEE286 pKa = 5.31GDD288 pKa = 3.43VTLTFDD294 pKa = 4.18RR295 pKa = 11.84VQEE298 pKa = 4.06AKK300 pKa = 10.36AAPDD304 pKa = 3.3LAFRR308 pKa = 11.84DD309 pKa = 4.03IIKK312 pKa = 10.36LSRR315 pKa = 11.84PIYY318 pKa = 10.35SEE320 pKa = 4.21APGGRR325 pKa = 11.84VRR327 pKa = 11.84VSRR330 pKa = 11.84LGTKK334 pKa = 8.53GTIRR338 pKa = 11.84LRR340 pKa = 11.84SGTLIGGQTHH350 pKa = 6.1YY351 pKa = 10.82FRR353 pKa = 11.84DD354 pKa = 3.84LSSIGAPSLEE364 pKa = 4.03MSVLGQTSGEE374 pKa = 4.31STVVLGSDD382 pKa = 3.28SSIINGVGLEE392 pKa = 4.16GTVAFPEE399 pKa = 4.22EE400 pKa = 3.95QLLLDD405 pKa = 4.01EE406 pKa = 5.47LSEE409 pKa = 4.23NFQNSQLVFSDD420 pKa = 3.16SHH422 pKa = 6.4QSHH425 pKa = 7.19IIEE428 pKa = 4.55LPRR431 pKa = 11.84IAEE434 pKa = 4.41SPRR437 pKa = 11.84SSYY440 pKa = 11.04VIYY443 pKa = 8.8DD444 pKa = 3.8TADD447 pKa = 3.37STVVSHH453 pKa = 7.18KK454 pKa = 10.86GNEE457 pKa = 3.95NYY459 pKa = 10.28FISTLTPPDD468 pKa = 4.05TEE470 pKa = 5.23PDD472 pKa = 3.91TEE474 pKa = 4.26PQTPDD479 pKa = 3.01EE480 pKa = 4.47TPRR483 pKa = 11.84APRR486 pKa = 11.84RR487 pKa = 11.84PLDD490 pKa = 4.3DD491 pKa = 4.25SDD493 pKa = 5.1GLTFDD498 pKa = 4.33LHH500 pKa = 6.56PGLLRR505 pKa = 11.84RR506 pKa = 11.84KK507 pKa = 8.63KK508 pKa = 10.4KK509 pKa = 9.51RR510 pKa = 11.84VRR512 pKa = 11.84YY513 pKa = 9.54YY514 pKa = 10.22IEE516 pKa = 3.8
Molecular weight: 55.06 kDa
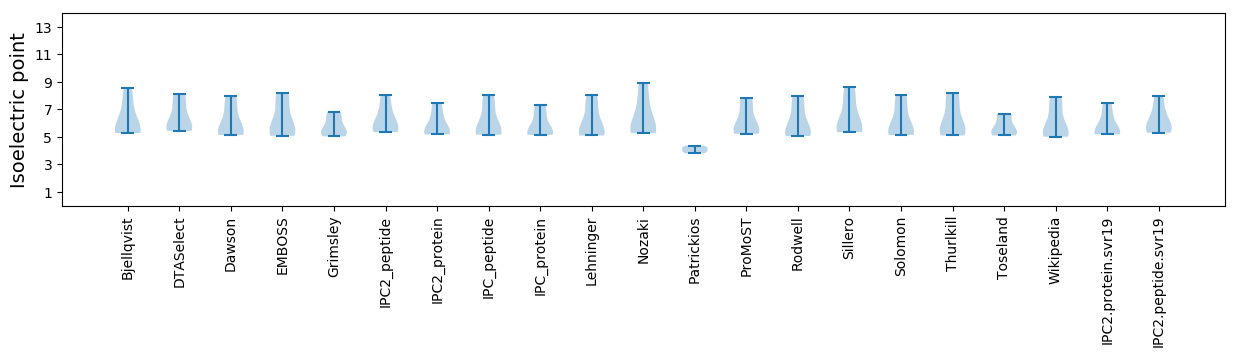
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2I8B2Q3|A0A2I8B2Q3_9PAPI Protein E7 OS=Leptonychotes weddellii papillomavirus 5 OX=2077306 GN=E7 PE=3 SV=1
MM1 pKa = 7.89EE2 pKa = 5.27KK3 pKa = 10.31PQTLRR8 pKa = 11.84ALAASLGEE16 pKa = 3.86PLQRR20 pKa = 11.84ILVQCVFCGAPMTWPDD36 pKa = 3.41KK37 pKa = 10.98VAFEE41 pKa = 4.32NKK43 pKa = 9.13CLSVVWCRR51 pKa = 11.84EE52 pKa = 3.82RR53 pKa = 11.84FFGACVSCTNVRR65 pKa = 11.84SYY67 pKa = 11.33WDD69 pKa = 3.3VLNNRR74 pKa = 11.84GPTLEE79 pKa = 4.04ATGVEE84 pKa = 4.36SLFNKK89 pKa = 9.41PLSDD93 pKa = 3.24IPVRR97 pKa = 11.84CMYY100 pKa = 10.58CLAQLTVLEE109 pKa = 5.16KK110 pKa = 10.34IACAVNEE117 pKa = 4.2RR118 pKa = 11.84PFVLVRR124 pKa = 11.84KK125 pKa = 9.44LFRR128 pKa = 11.84NTCSEE133 pKa = 3.98CTQYY137 pKa = 11.38DD138 pKa = 3.55RR139 pKa = 11.84SS140 pKa = 3.74
MM1 pKa = 7.89EE2 pKa = 5.27KK3 pKa = 10.31PQTLRR8 pKa = 11.84ALAASLGEE16 pKa = 3.86PLQRR20 pKa = 11.84ILVQCVFCGAPMTWPDD36 pKa = 3.41KK37 pKa = 10.98VAFEE41 pKa = 4.32NKK43 pKa = 9.13CLSVVWCRR51 pKa = 11.84EE52 pKa = 3.82RR53 pKa = 11.84FFGACVSCTNVRR65 pKa = 11.84SYY67 pKa = 11.33WDD69 pKa = 3.3VLNNRR74 pKa = 11.84GPTLEE79 pKa = 4.04ATGVEE84 pKa = 4.36SLFNKK89 pKa = 9.41PLSDD93 pKa = 3.24IPVRR97 pKa = 11.84CMYY100 pKa = 10.58CLAQLTVLEE109 pKa = 5.16KK110 pKa = 10.34IACAVNEE117 pKa = 4.2RR118 pKa = 11.84PFVLVRR124 pKa = 11.84KK125 pKa = 9.44LFRR128 pKa = 11.84NTCSEE133 pKa = 3.98CTQYY137 pKa = 11.38DD138 pKa = 3.55RR139 pKa = 11.84SS140 pKa = 3.74
Molecular weight: 15.88 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2416 |
121 |
601 |
402.7 |
44.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.381 ± 0.584 | 2.442 ± 0.707 |
6.167 ± 0.327 | 6.871 ± 0.517 |
4.553 ± 0.57 | 8.609 ± 1.142 |
1.987 ± 0.211 | 3.684 ± 0.265 |
5.05 ± 0.799 | 8.609 ± 0.445 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.78 ± 0.326 | 3.56 ± 0.521 |
6.747 ± 0.658 | 3.518 ± 0.484 |
6.291 ± 0.714 | 8.195 ± 0.432 |
6.25 ± 0.534 | 6.167 ± 0.665 |
1.242 ± 0.291 | 2.897 ± 0.385 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
