
Avian paramyxovirus UPO216
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Avulavirinae; Orthoavulavirus; Avian orthoavulavirus 16
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
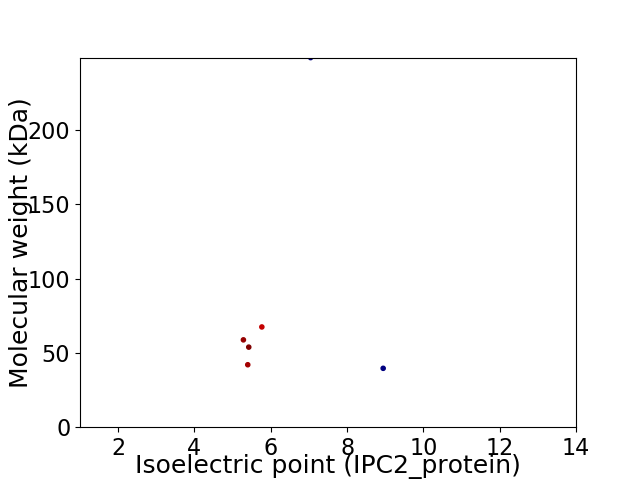
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Z1G744|A0A1Z1G744_9MONO Nucleocapsid OS=Avian paramyxovirus UPO216 OX=2006690 GN=NP PE=3 SV=1
MM1 pKa = 7.7IFTMYY6 pKa = 10.3HH7 pKa = 4.07VTVLLLLSLLTLPLGIQLARR27 pKa = 11.84ASIDD31 pKa = 3.23GRR33 pKa = 11.84QLAAAGIVVTGEE45 pKa = 3.95KK46 pKa = 10.28AINLYY51 pKa = 9.85TSSQTGTIVVKK62 pKa = 10.29LLPNVPQGRR71 pKa = 11.84EE72 pKa = 3.49ACMRR76 pKa = 11.84DD77 pKa = 3.35PLTSYY82 pKa = 11.45NKK84 pKa = 9.43TLTSLLSPLGEE95 pKa = 4.44AIRR98 pKa = 11.84RR99 pKa = 11.84IHH101 pKa = 6.94EE102 pKa = 4.49STTEE106 pKa = 3.89TAGLVQARR114 pKa = 11.84LVGAIIGSVALGVATSAQITAAAALIQANKK144 pKa = 9.49NAEE147 pKa = 4.37NILKK151 pKa = 10.2LKK153 pKa = 10.52QSIAATNEE161 pKa = 3.69AVHH164 pKa = 6.12EE165 pKa = 4.43VTDD168 pKa = 4.28GLSQLAVAVGKK179 pKa = 8.56MQDD182 pKa = 4.1FINTQFNNTAQEE194 pKa = 3.9IDD196 pKa = 4.56CIRR199 pKa = 11.84ISQQLGVEE207 pKa = 4.12LNLYY211 pKa = 8.46LTEE214 pKa = 4.14LTTVFGPQITSPALSPLSIQALYY237 pKa = 10.83NLAGGNLDD245 pKa = 3.61VLLSKK250 pKa = 10.64IGVGNNQLSALISSGLISGSPILYY274 pKa = 10.22DD275 pKa = 3.89SQTQLLGIQVTLPSVSSLNNMRR297 pKa = 11.84AIFLEE302 pKa = 4.17TLSVSTDD309 pKa = 2.87KK310 pKa = 11.47GFAAALIPKK319 pKa = 9.39VVTTVGTVTEE329 pKa = 4.37EE330 pKa = 4.68LDD332 pKa = 3.11TSYY335 pKa = 11.1CIEE338 pKa = 3.86TDD340 pKa = 2.54IDD342 pKa = 4.24LFCTRR347 pKa = 11.84IVTFPMSPGIYY358 pKa = 10.03ACLNGNTSEE367 pKa = 4.19CMYY370 pKa = 10.92SKK372 pKa = 9.2TQGALTTPYY381 pKa = 9.5MSVKK385 pKa = 10.27GSIVANCKK393 pKa = 7.6MTTCRR398 pKa = 11.84CADD401 pKa = 3.65PASIISQNYY410 pKa = 8.04GEE412 pKa = 4.78AVSLIDD418 pKa = 4.09SSVCRR423 pKa = 11.84VITLDD428 pKa = 3.45GVTLRR433 pKa = 11.84LSGSFDD439 pKa = 2.97STYY442 pKa = 10.95QKK444 pKa = 11.11NITIRR449 pKa = 11.84DD450 pKa = 3.69SQVIITGSLDD460 pKa = 2.88ISTEE464 pKa = 3.79LGNVNNSINNALDD477 pKa = 3.98KK478 pKa = 10.92IEE480 pKa = 4.61EE481 pKa = 4.35SNQILEE487 pKa = 4.41SVNVSLTSTNALIVYY502 pKa = 7.62IICTALALICGITGLILSCYY522 pKa = 9.89IMYY525 pKa = 11.01KK526 pKa = 9.36MRR528 pKa = 11.84SQQKK532 pKa = 7.67TLMWLGNNTLDD543 pKa = 4.73QMRR546 pKa = 11.84AQTKK550 pKa = 6.77MM551 pKa = 3.14
MM1 pKa = 7.7IFTMYY6 pKa = 10.3HH7 pKa = 4.07VTVLLLLSLLTLPLGIQLARR27 pKa = 11.84ASIDD31 pKa = 3.23GRR33 pKa = 11.84QLAAAGIVVTGEE45 pKa = 3.95KK46 pKa = 10.28AINLYY51 pKa = 9.85TSSQTGTIVVKK62 pKa = 10.29LLPNVPQGRR71 pKa = 11.84EE72 pKa = 3.49ACMRR76 pKa = 11.84DD77 pKa = 3.35PLTSYY82 pKa = 11.45NKK84 pKa = 9.43TLTSLLSPLGEE95 pKa = 4.44AIRR98 pKa = 11.84RR99 pKa = 11.84IHH101 pKa = 6.94EE102 pKa = 4.49STTEE106 pKa = 3.89TAGLVQARR114 pKa = 11.84LVGAIIGSVALGVATSAQITAAAALIQANKK144 pKa = 9.49NAEE147 pKa = 4.37NILKK151 pKa = 10.2LKK153 pKa = 10.52QSIAATNEE161 pKa = 3.69AVHH164 pKa = 6.12EE165 pKa = 4.43VTDD168 pKa = 4.28GLSQLAVAVGKK179 pKa = 8.56MQDD182 pKa = 4.1FINTQFNNTAQEE194 pKa = 3.9IDD196 pKa = 4.56CIRR199 pKa = 11.84ISQQLGVEE207 pKa = 4.12LNLYY211 pKa = 8.46LTEE214 pKa = 4.14LTTVFGPQITSPALSPLSIQALYY237 pKa = 10.83NLAGGNLDD245 pKa = 3.61VLLSKK250 pKa = 10.64IGVGNNQLSALISSGLISGSPILYY274 pKa = 10.22DD275 pKa = 3.89SQTQLLGIQVTLPSVSSLNNMRR297 pKa = 11.84AIFLEE302 pKa = 4.17TLSVSTDD309 pKa = 2.87KK310 pKa = 11.47GFAAALIPKK319 pKa = 9.39VVTTVGTVTEE329 pKa = 4.37EE330 pKa = 4.68LDD332 pKa = 3.11TSYY335 pKa = 11.1CIEE338 pKa = 3.86TDD340 pKa = 2.54IDD342 pKa = 4.24LFCTRR347 pKa = 11.84IVTFPMSPGIYY358 pKa = 10.03ACLNGNTSEE367 pKa = 4.19CMYY370 pKa = 10.92SKK372 pKa = 9.2TQGALTTPYY381 pKa = 9.5MSVKK385 pKa = 10.27GSIVANCKK393 pKa = 7.6MTTCRR398 pKa = 11.84CADD401 pKa = 3.65PASIISQNYY410 pKa = 8.04GEE412 pKa = 4.78AVSLIDD418 pKa = 4.09SSVCRR423 pKa = 11.84VITLDD428 pKa = 3.45GVTLRR433 pKa = 11.84LSGSFDD439 pKa = 2.97STYY442 pKa = 10.95QKK444 pKa = 11.11NITIRR449 pKa = 11.84DD450 pKa = 3.69SQVIITGSLDD460 pKa = 2.88ISTEE464 pKa = 3.79LGNVNNSINNALDD477 pKa = 3.98KK478 pKa = 10.92IEE480 pKa = 4.61EE481 pKa = 4.35SNQILEE487 pKa = 4.41SVNVSLTSTNALIVYY502 pKa = 7.62IICTALALICGITGLILSCYY522 pKa = 9.89IMYY525 pKa = 11.01KK526 pKa = 9.36MRR528 pKa = 11.84SQQKK532 pKa = 7.67TLMWLGNNTLDD543 pKa = 4.73QMRR546 pKa = 11.84AQTKK550 pKa = 6.77MM551 pKa = 3.14
Molecular weight: 58.79 kDa
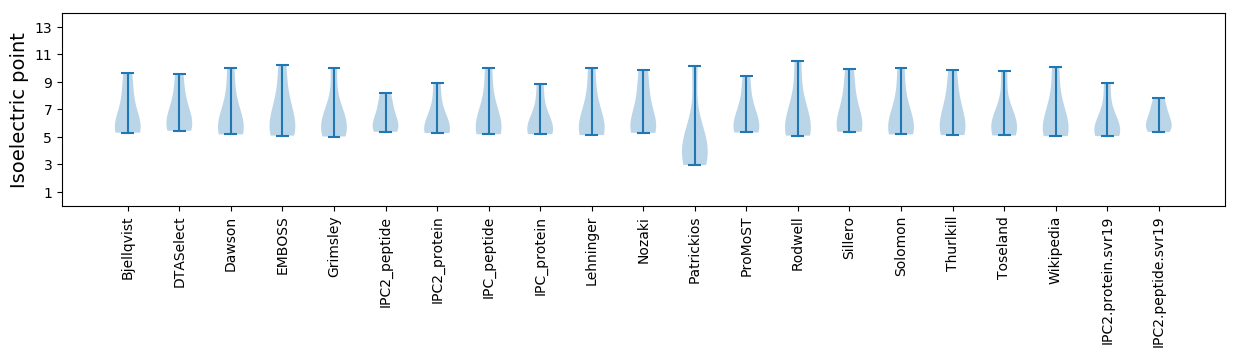
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Z1G749|A0A1Z1G749_9MONO Matrix protein OS=Avian paramyxovirus UPO216 OX=2006690 GN=M PE=4 SV=1
MM1 pKa = 8.22DD2 pKa = 3.47STRR5 pKa = 11.84TIGIHH10 pKa = 6.85FDD12 pKa = 3.35PTLPASSLLAFPIVLQEE29 pKa = 4.06TGDD32 pKa = 3.69GKK34 pKa = 11.03KK35 pKa = 10.19QITPQYY41 pKa = 9.77RR42 pKa = 11.84IQKK45 pKa = 8.68LDD47 pKa = 2.89SWTEE51 pKa = 3.94SKK53 pKa = 10.89DD54 pKa = 3.28DD55 pKa = 3.62SLFITTYY62 pKa = 10.76GFIFRR67 pKa = 11.84VGGGGSNMGTIEE79 pKa = 4.01GTPTRR84 pKa = 11.84EE85 pKa = 3.98LLSAAMLCIGSVSNTTDD102 pKa = 3.06PVEE105 pKa = 3.92IARR108 pKa = 11.84ACLSLMITCKK118 pKa = 10.39KK119 pKa = 10.38SATNTEE125 pKa = 3.73RR126 pKa = 11.84MIFSIVQAPQILQSCKK142 pKa = 9.66VVANKK147 pKa = 9.64YY148 pKa = 10.07ASVSAVKK155 pKa = 10.03HH156 pKa = 4.56VKK158 pKa = 10.37APEE161 pKa = 4.02KK162 pKa = 10.96VPGDD166 pKa = 3.3GTLEE170 pKa = 4.02YY171 pKa = 10.52KK172 pKa = 10.99VNFVSLTVVPKK183 pKa = 10.08RR184 pKa = 11.84DD185 pKa = 3.37VYY187 pKa = 10.77RR188 pKa = 11.84IPSTALRR195 pKa = 11.84VSGPSLYY202 pKa = 10.95NLALNVIIAVEE213 pKa = 4.02VDD215 pKa = 3.78DD216 pKa = 5.54KK217 pKa = 11.99SPLTRR222 pKa = 11.84SLTRR226 pKa = 11.84TEE228 pKa = 4.74EE229 pKa = 4.4GFCANLFLHH238 pKa = 6.97IGLMSTVDD246 pKa = 3.43KK247 pKa = 10.63KK248 pKa = 11.2GKK250 pKa = 10.07KK251 pKa = 9.39ISFDD255 pKa = 3.06KK256 pKa = 11.14LEE258 pKa = 4.17RR259 pKa = 11.84KK260 pKa = 9.0IRR262 pKa = 11.84RR263 pKa = 11.84LDD265 pKa = 3.57LSVGLSDD272 pKa = 4.26VLGPSILIKK281 pKa = 10.78ARR283 pKa = 11.84GARR286 pKa = 11.84TRR288 pKa = 11.84LMSPFFSSSGTACYY302 pKa = 9.57PIANASPQVAKK313 pKa = 10.65ILWSQSASLRR323 pKa = 11.84SVKK326 pKa = 10.13IVIQAGTQKK335 pKa = 10.81AIAVTADD342 pKa = 3.47HH343 pKa = 6.65EE344 pKa = 4.93VMSTKK349 pKa = 10.43LEE351 pKa = 4.19KK352 pKa = 10.79NHH354 pKa = 6.76TISKK358 pKa = 9.32FNPFKK363 pKa = 10.85KK364 pKa = 10.49
MM1 pKa = 8.22DD2 pKa = 3.47STRR5 pKa = 11.84TIGIHH10 pKa = 6.85FDD12 pKa = 3.35PTLPASSLLAFPIVLQEE29 pKa = 4.06TGDD32 pKa = 3.69GKK34 pKa = 11.03KK35 pKa = 10.19QITPQYY41 pKa = 9.77RR42 pKa = 11.84IQKK45 pKa = 8.68LDD47 pKa = 2.89SWTEE51 pKa = 3.94SKK53 pKa = 10.89DD54 pKa = 3.28DD55 pKa = 3.62SLFITTYY62 pKa = 10.76GFIFRR67 pKa = 11.84VGGGGSNMGTIEE79 pKa = 4.01GTPTRR84 pKa = 11.84EE85 pKa = 3.98LLSAAMLCIGSVSNTTDD102 pKa = 3.06PVEE105 pKa = 3.92IARR108 pKa = 11.84ACLSLMITCKK118 pKa = 10.39KK119 pKa = 10.38SATNTEE125 pKa = 3.73RR126 pKa = 11.84MIFSIVQAPQILQSCKK142 pKa = 9.66VVANKK147 pKa = 9.64YY148 pKa = 10.07ASVSAVKK155 pKa = 10.03HH156 pKa = 4.56VKK158 pKa = 10.37APEE161 pKa = 4.02KK162 pKa = 10.96VPGDD166 pKa = 3.3GTLEE170 pKa = 4.02YY171 pKa = 10.52KK172 pKa = 10.99VNFVSLTVVPKK183 pKa = 10.08RR184 pKa = 11.84DD185 pKa = 3.37VYY187 pKa = 10.77RR188 pKa = 11.84IPSTALRR195 pKa = 11.84VSGPSLYY202 pKa = 10.95NLALNVIIAVEE213 pKa = 4.02VDD215 pKa = 3.78DD216 pKa = 5.54KK217 pKa = 11.99SPLTRR222 pKa = 11.84SLTRR226 pKa = 11.84TEE228 pKa = 4.74EE229 pKa = 4.4GFCANLFLHH238 pKa = 6.97IGLMSTVDD246 pKa = 3.43KK247 pKa = 10.63KK248 pKa = 11.2GKK250 pKa = 10.07KK251 pKa = 9.39ISFDD255 pKa = 3.06KK256 pKa = 11.14LEE258 pKa = 4.17RR259 pKa = 11.84KK260 pKa = 9.0IRR262 pKa = 11.84RR263 pKa = 11.84LDD265 pKa = 3.57LSVGLSDD272 pKa = 4.26VLGPSILIKK281 pKa = 10.78ARR283 pKa = 11.84GARR286 pKa = 11.84TRR288 pKa = 11.84LMSPFFSSSGTACYY302 pKa = 9.57PIANASPQVAKK313 pKa = 10.65ILWSQSASLRR323 pKa = 11.84SVKK326 pKa = 10.13IVIQAGTQKK335 pKa = 10.81AIAVTADD342 pKa = 3.47HH343 pKa = 6.65EE344 pKa = 4.93VMSTKK349 pKa = 10.43LEE351 pKa = 4.19KK352 pKa = 10.79NHH354 pKa = 6.76TISKK358 pKa = 9.32FNPFKK363 pKa = 10.85KK364 pKa = 10.49
Molecular weight: 39.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4625 |
364 |
2202 |
770.8 |
85.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.33 ± 0.755 | 1.795 ± 0.202 |
4.865 ± 0.285 | 4.995 ± 0.382 |
3.395 ± 0.598 | 5.946 ± 0.599 |
1.881 ± 0.441 | 6.746 ± 0.456 |
4.757 ± 0.427 | 10.551 ± 1.002 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.832 ± 0.252 | 4.67 ± 0.294 |
4.8 ± 0.566 | 4.173 ± 0.498 |
5.059 ± 0.531 | 9.016 ± 0.392 |
7.178 ± 0.726 | 6.119 ± 0.253 |
0.692 ± 0.131 | 3.2 ± 0.452 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
