
Streptomyces sp. PT12
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; unclassified Streptomyces
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
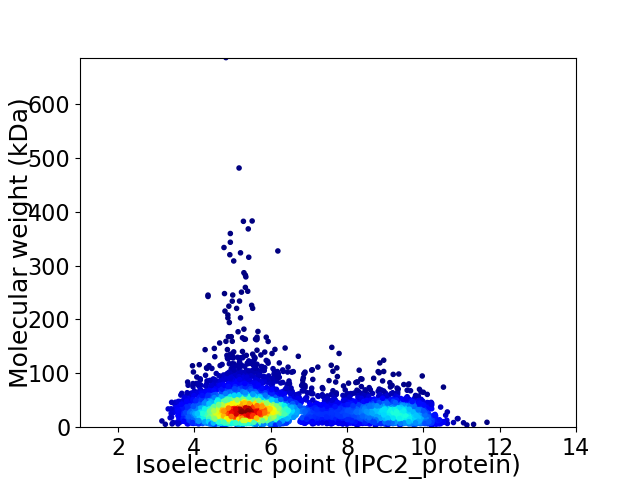
Virtual 2D-PAGE plot for 5724 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A365ZWD7|A0A365ZWD7_9ACTN GNAT family N-acetyltransferase OS=Streptomyces sp. PT12 OX=1510197 GN=DEH69_03005 PE=4 SV=1
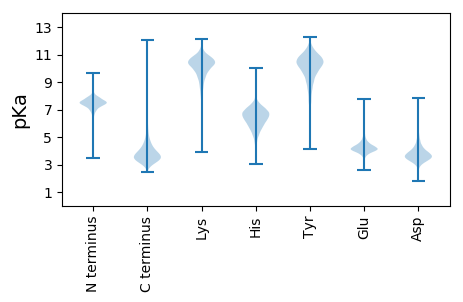
PP1 pKa = 6.84GASVSTGANFTYY13 pKa = 10.34SGANAAPTAFSVNGVVCGDD32 pKa = 4.21DD33 pKa = 4.13GGPGPDD39 pKa = 4.85PDD41 pKa = 5.47PDD43 pKa = 4.97PDD45 pKa = 4.82PDD47 pKa = 4.05PPGDD51 pKa = 3.66RR52 pKa = 11.84VDD54 pKa = 4.65NPYY57 pKa = 10.82EE58 pKa = 4.09DD59 pKa = 3.46AGVYY63 pKa = 9.63VNPDD67 pKa = 2.61WSANAAAEE75 pKa = 4.21PGGDD79 pKa = 4.19AIADD83 pKa = 3.7EE84 pKa = 4.67PTAVWLDD91 pKa = 3.35RR92 pKa = 11.84RR93 pKa = 11.84AAITGGSAGNSTGLRR108 pKa = 11.84EE109 pKa = 4.31HH110 pKa = 7.37LDD112 pKa = 3.3NALIQAEE119 pKa = 4.16EE120 pKa = 3.87HH121 pKa = 7.19DD122 pKa = 3.82NFVFQVVVYY131 pKa = 8.6NLPGRR136 pKa = 11.84DD137 pKa = 3.48CAALASNGEE146 pKa = 4.12LAADD150 pKa = 4.45EE151 pKa = 3.99IDD153 pKa = 3.72VYY155 pKa = 11.07KK156 pKa = 10.94EE157 pKa = 3.77EE158 pKa = 5.06FIDD161 pKa = 5.08PIAEE165 pKa = 4.08ILGDD169 pKa = 3.59PAYY172 pKa = 10.66ADD174 pKa = 3.76LRR176 pKa = 11.84IVTIIEE182 pKa = 3.95IDD184 pKa = 3.68SLPNLITNVSPRR196 pKa = 11.84EE197 pKa = 4.02TATDD201 pKa = 3.16QCDD204 pKa = 3.17EE205 pKa = 4.03MLANGNYY212 pKa = 9.54VEE214 pKa = 4.41GVSYY218 pKa = 11.19ALDD221 pKa = 3.61TLGDD225 pKa = 3.48IPNVYY230 pKa = 10.54NYY232 pKa = 9.82IDD234 pKa = 3.8AGHH237 pKa = 6.33YY238 pKa = 10.09GWIGWDD244 pKa = 3.6DD245 pKa = 3.82NFGASAEE252 pKa = 4.44LFHH255 pKa = 6.54EE256 pKa = 4.56TATTGGASVDD266 pKa = 3.83DD267 pKa = 3.78VAGFIVNTANYY278 pKa = 8.33GPSVEE283 pKa = 4.08PHH285 pKa = 5.83YY286 pKa = 11.34SIDD289 pKa = 3.51DD290 pKa = 3.9TVGGRR295 pKa = 11.84SVRR298 pKa = 11.84EE299 pKa = 4.18SEE301 pKa = 3.85WVDD304 pKa = 2.72WNRR307 pKa = 11.84YY308 pKa = 8.3IDD310 pKa = 3.78NQSFALAFRR319 pKa = 11.84DD320 pKa = 3.67EE321 pKa = 4.52LVSQGFNDD329 pKa = 4.48EE330 pKa = 3.89IGMLIDD336 pKa = 3.57TSRR339 pKa = 11.84NGWGGPDD346 pKa = 3.52RR347 pKa = 11.84PTGPGPTTDD356 pKa = 2.68VNAYY360 pKa = 10.07VEE362 pKa = 4.39GSRR365 pKa = 11.84VDD367 pKa = 4.12GRR369 pKa = 11.84IQTGNWCNQSGAGLGEE385 pKa = 4.66RR386 pKa = 11.84PTASPVPNIDD396 pKa = 2.65AYY398 pKa = 10.94AWIKK402 pKa = 10.43PPGEE406 pKa = 4.03SDD408 pKa = 3.31GSSEE412 pKa = 5.19EE413 pKa = 4.0IPNDD417 pKa = 3.14EE418 pKa = 4.17GKK420 pKa = 11.07GFDD423 pKa = 3.72RR424 pKa = 11.84MCDD427 pKa = 3.22PTYY430 pKa = 10.84EE431 pKa = 4.29GNPRR435 pKa = 11.84NNNNMSGALPDD446 pKa = 4.3APLSGHH452 pKa = 6.02WFSAQFQEE460 pKa = 4.77LLANAYY466 pKa = 8.53PPLL469 pKa = 5.25
PP1 pKa = 6.84GASVSTGANFTYY13 pKa = 10.34SGANAAPTAFSVNGVVCGDD32 pKa = 4.21DD33 pKa = 4.13GGPGPDD39 pKa = 4.85PDD41 pKa = 5.47PDD43 pKa = 4.97PDD45 pKa = 4.82PDD47 pKa = 4.05PPGDD51 pKa = 3.66RR52 pKa = 11.84VDD54 pKa = 4.65NPYY57 pKa = 10.82EE58 pKa = 4.09DD59 pKa = 3.46AGVYY63 pKa = 9.63VNPDD67 pKa = 2.61WSANAAAEE75 pKa = 4.21PGGDD79 pKa = 4.19AIADD83 pKa = 3.7EE84 pKa = 4.67PTAVWLDD91 pKa = 3.35RR92 pKa = 11.84RR93 pKa = 11.84AAITGGSAGNSTGLRR108 pKa = 11.84EE109 pKa = 4.31HH110 pKa = 7.37LDD112 pKa = 3.3NALIQAEE119 pKa = 4.16EE120 pKa = 3.87HH121 pKa = 7.19DD122 pKa = 3.82NFVFQVVVYY131 pKa = 8.6NLPGRR136 pKa = 11.84DD137 pKa = 3.48CAALASNGEE146 pKa = 4.12LAADD150 pKa = 4.45EE151 pKa = 3.99IDD153 pKa = 3.72VYY155 pKa = 11.07KK156 pKa = 10.94EE157 pKa = 3.77EE158 pKa = 5.06FIDD161 pKa = 5.08PIAEE165 pKa = 4.08ILGDD169 pKa = 3.59PAYY172 pKa = 10.66ADD174 pKa = 3.76LRR176 pKa = 11.84IVTIIEE182 pKa = 3.95IDD184 pKa = 3.68SLPNLITNVSPRR196 pKa = 11.84EE197 pKa = 4.02TATDD201 pKa = 3.16QCDD204 pKa = 3.17EE205 pKa = 4.03MLANGNYY212 pKa = 9.54VEE214 pKa = 4.41GVSYY218 pKa = 11.19ALDD221 pKa = 3.61TLGDD225 pKa = 3.48IPNVYY230 pKa = 10.54NYY232 pKa = 9.82IDD234 pKa = 3.8AGHH237 pKa = 6.33YY238 pKa = 10.09GWIGWDD244 pKa = 3.6DD245 pKa = 3.82NFGASAEE252 pKa = 4.44LFHH255 pKa = 6.54EE256 pKa = 4.56TATTGGASVDD266 pKa = 3.83DD267 pKa = 3.78VAGFIVNTANYY278 pKa = 8.33GPSVEE283 pKa = 4.08PHH285 pKa = 5.83YY286 pKa = 11.34SIDD289 pKa = 3.51DD290 pKa = 3.9TVGGRR295 pKa = 11.84SVRR298 pKa = 11.84EE299 pKa = 4.18SEE301 pKa = 3.85WVDD304 pKa = 2.72WNRR307 pKa = 11.84YY308 pKa = 8.3IDD310 pKa = 3.78NQSFALAFRR319 pKa = 11.84DD320 pKa = 3.67EE321 pKa = 4.52LVSQGFNDD329 pKa = 4.48EE330 pKa = 3.89IGMLIDD336 pKa = 3.57TSRR339 pKa = 11.84NGWGGPDD346 pKa = 3.52RR347 pKa = 11.84PTGPGPTTDD356 pKa = 2.68VNAYY360 pKa = 10.07VEE362 pKa = 4.39GSRR365 pKa = 11.84VDD367 pKa = 4.12GRR369 pKa = 11.84IQTGNWCNQSGAGLGEE385 pKa = 4.66RR386 pKa = 11.84PTASPVPNIDD396 pKa = 2.65AYY398 pKa = 10.94AWIKK402 pKa = 10.43PPGEE406 pKa = 4.03SDD408 pKa = 3.31GSSEE412 pKa = 5.19EE413 pKa = 4.0IPNDD417 pKa = 3.14EE418 pKa = 4.17GKK420 pKa = 11.07GFDD423 pKa = 3.72RR424 pKa = 11.84MCDD427 pKa = 3.22PTYY430 pKa = 10.84EE431 pKa = 4.29GNPRR435 pKa = 11.84NNNNMSGALPDD446 pKa = 4.3APLSGHH452 pKa = 6.02WFSAQFQEE460 pKa = 4.77LLANAYY466 pKa = 8.53PPLL469 pKa = 5.25
Molecular weight: 49.88 kDa
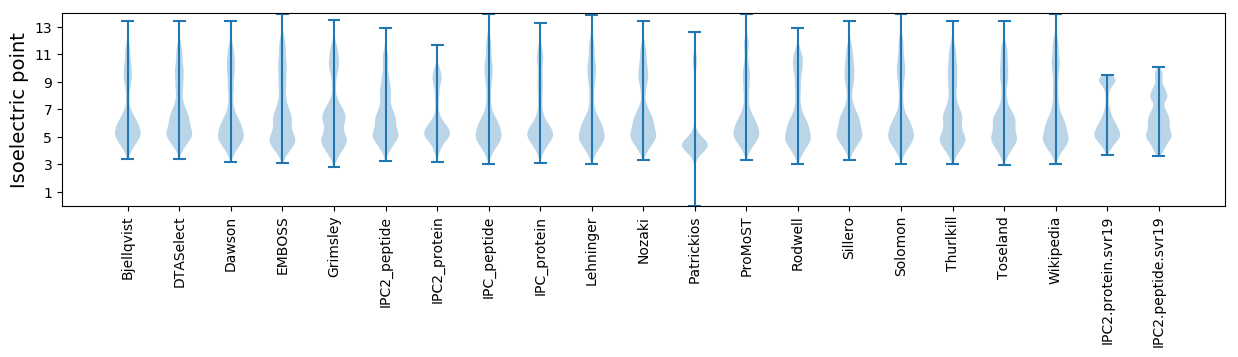
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A365YS73|A0A365YS73_9ACTN Site-specific integrase OS=Streptomyces sp. PT12 OX=1510197 GN=DEH69_28125 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AVIANRR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.82GRR40 pKa = 11.84SRR42 pKa = 11.84LSAA45 pKa = 3.74
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AVIANRR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.82GRR40 pKa = 11.84SRR42 pKa = 11.84LSAA45 pKa = 3.74
Molecular weight: 5.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1969865 |
22 |
6549 |
344.1 |
36.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.433 ± 0.061 | 0.77 ± 0.009 |
6.145 ± 0.029 | 6.032 ± 0.033 |
2.659 ± 0.017 | 9.772 ± 0.035 |
2.377 ± 0.016 | 3.033 ± 0.02 |
1.362 ± 0.021 | 10.462 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.597 ± 0.013 | 1.575 ± 0.017 |
6.448 ± 0.034 | 2.425 ± 0.018 |
8.754 ± 0.036 | 4.735 ± 0.022 |
5.8 ± 0.027 | 8.225 ± 0.027 |
1.535 ± 0.016 | 1.863 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
