
Candidatus Halobonum tyrrellensis G22
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Haloferacales; Halorubraceae; Candidatus Halobonum; Candidatus Halobonum tyrrellensis
Average proteome isoelectric point is 4.95
Get precalculated fractions of proteins
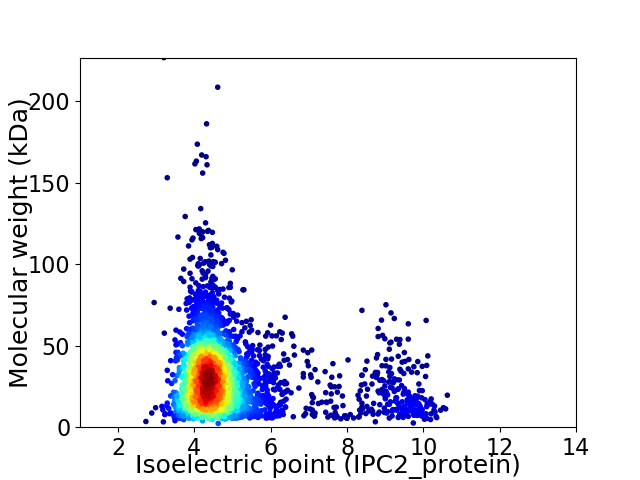
Virtual 2D-PAGE plot for 3445 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|V4HBA0|V4HBA0_9EURY Uncharacterized protein OS=Candidatus Halobonum tyrrellensis G22 OX=1324957 GN=K933_14468 PE=4 SV=1
MM1 pKa = 7.69SDD3 pKa = 3.27YY4 pKa = 11.4AKK6 pKa = 10.71DD7 pKa = 3.81VLVSADD13 pKa = 3.23WVEE16 pKa = 3.85EE17 pKa = 3.91HH18 pKa = 7.19LDD20 pKa = 3.7EE21 pKa = 5.3FQSDD25 pKa = 3.57DD26 pKa = 3.48DD27 pKa = 4.17EE28 pKa = 4.93YY29 pKa = 11.73RR30 pKa = 11.84LVEE33 pKa = 3.98VDD35 pKa = 4.17VDD37 pKa = 3.84TEE39 pKa = 4.62AYY41 pKa = 10.29DD42 pKa = 3.54SGHH45 pKa = 6.64APGAIGFNWEE55 pKa = 4.26TQLQDD60 pKa = 3.03QTRR63 pKa = 11.84RR64 pKa = 11.84DD65 pKa = 3.71LLTTEE70 pKa = 5.1DD71 pKa = 4.87FEE73 pKa = 5.96DD74 pKa = 4.18LNASHH79 pKa = 7.27GISNDD84 pKa = 3.2STVVIYY90 pKa = 10.83GDD92 pKa = 3.53SSNWFAAYY100 pKa = 8.43TYY102 pKa = 8.79WQYY105 pKa = 11.8KK106 pKa = 9.41YY107 pKa = 10.41YY108 pKa = 10.98GHH110 pKa = 7.78DD111 pKa = 3.99DD112 pKa = 3.38VRR114 pKa = 11.84LMDD117 pKa = 4.23GGRR120 pKa = 11.84EE121 pKa = 3.84YY122 pKa = 10.27WVDD125 pKa = 2.94NDD127 pKa = 3.9YY128 pKa = 8.15PTTDD132 pKa = 4.03EE133 pKa = 4.57EE134 pKa = 4.64PSFPEE139 pKa = 3.89TEE141 pKa = 4.19YY142 pKa = 11.13EE143 pKa = 3.82AAEE146 pKa = 4.14PDD148 pKa = 3.45EE149 pKa = 4.97SIRR152 pKa = 11.84AYY154 pKa = 10.55RR155 pKa = 11.84EE156 pKa = 3.7QVDD159 pKa = 3.44AAIDD163 pKa = 3.6AGVPLVDD170 pKa = 3.35VRR172 pKa = 11.84SPEE175 pKa = 3.94EE176 pKa = 3.64FRR178 pKa = 11.84GEE180 pKa = 3.65ILAPPGLQEE189 pKa = 3.56TAQRR193 pKa = 11.84GGHH196 pKa = 6.15IPGASNISWASTVNEE211 pKa = 4.56DD212 pKa = 4.16GTFKK216 pKa = 10.42TADD219 pKa = 3.71EE220 pKa = 4.16LQEE223 pKa = 4.33LYY225 pKa = 10.88EE226 pKa = 4.19GVIDD230 pKa = 5.16AGDD233 pKa = 3.91DD234 pKa = 3.82EE235 pKa = 4.64IVAYY239 pKa = 10.34CRR241 pKa = 11.84IGEE244 pKa = 4.16RR245 pKa = 11.84SSIAWFALHH254 pKa = 6.86EE255 pKa = 4.66LLGQDD260 pKa = 2.8QTVNYY265 pKa = 9.6DD266 pKa = 3.59GSWTEE271 pKa = 3.62WGNLVGAPVEE281 pKa = 4.36TGDD284 pKa = 4.99AEE286 pKa = 4.12
MM1 pKa = 7.69SDD3 pKa = 3.27YY4 pKa = 11.4AKK6 pKa = 10.71DD7 pKa = 3.81VLVSADD13 pKa = 3.23WVEE16 pKa = 3.85EE17 pKa = 3.91HH18 pKa = 7.19LDD20 pKa = 3.7EE21 pKa = 5.3FQSDD25 pKa = 3.57DD26 pKa = 3.48DD27 pKa = 4.17EE28 pKa = 4.93YY29 pKa = 11.73RR30 pKa = 11.84LVEE33 pKa = 3.98VDD35 pKa = 4.17VDD37 pKa = 3.84TEE39 pKa = 4.62AYY41 pKa = 10.29DD42 pKa = 3.54SGHH45 pKa = 6.64APGAIGFNWEE55 pKa = 4.26TQLQDD60 pKa = 3.03QTRR63 pKa = 11.84RR64 pKa = 11.84DD65 pKa = 3.71LLTTEE70 pKa = 5.1DD71 pKa = 4.87FEE73 pKa = 5.96DD74 pKa = 4.18LNASHH79 pKa = 7.27GISNDD84 pKa = 3.2STVVIYY90 pKa = 10.83GDD92 pKa = 3.53SSNWFAAYY100 pKa = 8.43TYY102 pKa = 8.79WQYY105 pKa = 11.8KK106 pKa = 9.41YY107 pKa = 10.41YY108 pKa = 10.98GHH110 pKa = 7.78DD111 pKa = 3.99DD112 pKa = 3.38VRR114 pKa = 11.84LMDD117 pKa = 4.23GGRR120 pKa = 11.84EE121 pKa = 3.84YY122 pKa = 10.27WVDD125 pKa = 2.94NDD127 pKa = 3.9YY128 pKa = 8.15PTTDD132 pKa = 4.03EE133 pKa = 4.57EE134 pKa = 4.64PSFPEE139 pKa = 3.89TEE141 pKa = 4.19YY142 pKa = 11.13EE143 pKa = 3.82AAEE146 pKa = 4.14PDD148 pKa = 3.45EE149 pKa = 4.97SIRR152 pKa = 11.84AYY154 pKa = 10.55RR155 pKa = 11.84EE156 pKa = 3.7QVDD159 pKa = 3.44AAIDD163 pKa = 3.6AGVPLVDD170 pKa = 3.35VRR172 pKa = 11.84SPEE175 pKa = 3.94EE176 pKa = 3.64FRR178 pKa = 11.84GEE180 pKa = 3.65ILAPPGLQEE189 pKa = 3.56TAQRR193 pKa = 11.84GGHH196 pKa = 6.15IPGASNISWASTVNEE211 pKa = 4.56DD212 pKa = 4.16GTFKK216 pKa = 10.42TADD219 pKa = 3.71EE220 pKa = 4.16LQEE223 pKa = 4.33LYY225 pKa = 10.88EE226 pKa = 4.19GVIDD230 pKa = 5.16AGDD233 pKa = 3.91DD234 pKa = 3.82EE235 pKa = 4.64IVAYY239 pKa = 10.34CRR241 pKa = 11.84IGEE244 pKa = 4.16RR245 pKa = 11.84SSIAWFALHH254 pKa = 6.86EE255 pKa = 4.66LLGQDD260 pKa = 2.8QTVNYY265 pKa = 9.6DD266 pKa = 3.59GSWTEE271 pKa = 3.62WGNLVGAPVEE281 pKa = 4.36TGDD284 pKa = 4.99AEE286 pKa = 4.12
Molecular weight: 32.12 kDa
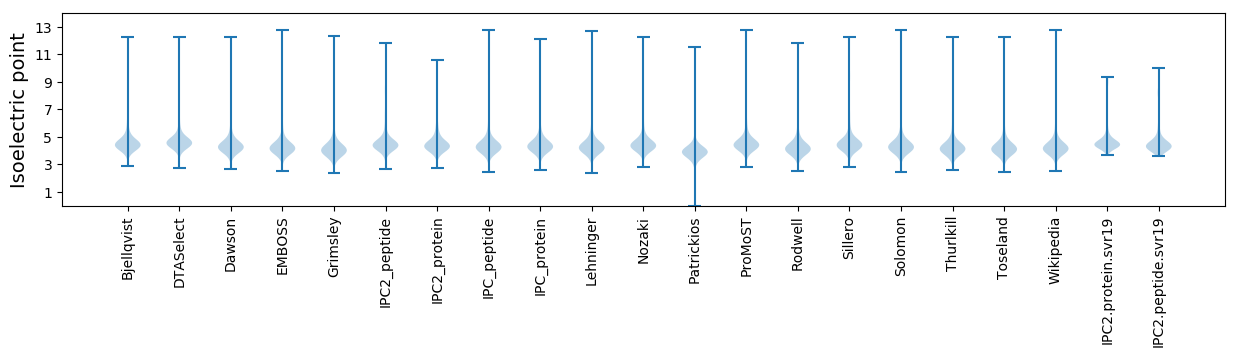
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|V4HGH7|V4HGH7_9EURY Putative hydrolase OS=Candidatus Halobonum tyrrellensis G22 OX=1324957 GN=K933_04351 PE=4 SV=1
MM1 pKa = 7.13YY2 pKa = 7.95RR3 pKa = 11.84TPHH6 pKa = 6.98DD7 pKa = 3.57EE8 pKa = 4.04HH9 pKa = 8.79DD10 pKa = 3.98GMKK13 pKa = 9.93VWLAADD19 pKa = 3.59EE20 pKa = 4.61ADD22 pKa = 4.95DD23 pKa = 4.01YY24 pKa = 11.77LALVEE29 pKa = 4.52DD30 pKa = 4.1TTHH33 pKa = 7.74RR34 pKa = 11.84IALSLGLRR42 pKa = 11.84CGLRR46 pKa = 11.84SAEE49 pKa = 4.12IVQTTPGDD57 pKa = 3.86LADD60 pKa = 4.75GPAGTMLRR68 pKa = 11.84VPDD71 pKa = 4.13GKK73 pKa = 10.49GGKK76 pKa = 8.93YY77 pKa = 9.52RR78 pKa = 11.84EE79 pKa = 4.34TPVPPEE85 pKa = 4.37LKK87 pKa = 8.34TTIQTVDD94 pKa = 3.11NVRR97 pKa = 11.84AEE99 pKa = 4.3PSSSPLVDD107 pKa = 3.18ASTRR111 pKa = 11.84TLRR114 pKa = 11.84RR115 pKa = 11.84WVRR118 pKa = 11.84KK119 pKa = 9.96SPLKK123 pKa = 10.13RR124 pKa = 11.84PKK126 pKa = 10.45SRR128 pKa = 11.84ATTAGGTSRR137 pKa = 11.84RR138 pKa = 11.84TTSGGHH144 pKa = 5.77GRR146 pKa = 11.84RR147 pKa = 11.84CSPEE151 pKa = 3.42PRR153 pKa = 11.84RR154 pKa = 11.84LTRR157 pKa = 11.84CSSVSGAGGRR167 pKa = 11.84TGNALRR173 pKa = 11.84ALPWRR178 pKa = 11.84LLSGGGTMRR187 pKa = 11.84GAA189 pKa = 3.8
MM1 pKa = 7.13YY2 pKa = 7.95RR3 pKa = 11.84TPHH6 pKa = 6.98DD7 pKa = 3.57EE8 pKa = 4.04HH9 pKa = 8.79DD10 pKa = 3.98GMKK13 pKa = 9.93VWLAADD19 pKa = 3.59EE20 pKa = 4.61ADD22 pKa = 4.95DD23 pKa = 4.01YY24 pKa = 11.77LALVEE29 pKa = 4.52DD30 pKa = 4.1TTHH33 pKa = 7.74RR34 pKa = 11.84IALSLGLRR42 pKa = 11.84CGLRR46 pKa = 11.84SAEE49 pKa = 4.12IVQTTPGDD57 pKa = 3.86LADD60 pKa = 4.75GPAGTMLRR68 pKa = 11.84VPDD71 pKa = 4.13GKK73 pKa = 10.49GGKK76 pKa = 8.93YY77 pKa = 9.52RR78 pKa = 11.84EE79 pKa = 4.34TPVPPEE85 pKa = 4.37LKK87 pKa = 8.34TTIQTVDD94 pKa = 3.11NVRR97 pKa = 11.84AEE99 pKa = 4.3PSSSPLVDD107 pKa = 3.18ASTRR111 pKa = 11.84TLRR114 pKa = 11.84RR115 pKa = 11.84WVRR118 pKa = 11.84KK119 pKa = 9.96SPLKK123 pKa = 10.13RR124 pKa = 11.84PKK126 pKa = 10.45SRR128 pKa = 11.84ATTAGGTSRR137 pKa = 11.84RR138 pKa = 11.84TTSGGHH144 pKa = 5.77GRR146 pKa = 11.84RR147 pKa = 11.84CSPEE151 pKa = 3.42PRR153 pKa = 11.84RR154 pKa = 11.84LTRR157 pKa = 11.84CSSVSGAGGRR167 pKa = 11.84TGNALRR173 pKa = 11.84ALPWRR178 pKa = 11.84LLSGGGTMRR187 pKa = 11.84GAA189 pKa = 3.8
Molecular weight: 20.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1030664 |
22 |
2237 |
299.2 |
32.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.37 ± 0.08 | 0.679 ± 0.015 |
8.921 ± 0.062 | 7.919 ± 0.06 |
3.274 ± 0.028 | 9.422 ± 0.047 |
1.857 ± 0.021 | 2.664 ± 0.038 |
1.366 ± 0.025 | 8.729 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.624 ± 0.019 | 2.046 ± 0.024 |
4.9 ± 0.026 | 1.885 ± 0.023 |
7.231 ± 0.05 | 5.262 ± 0.031 |
6.237 ± 0.037 | 9.789 ± 0.047 |
1.149 ± 0.018 | 2.676 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
