
Beihai tombus-like virus 9
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.28
Get precalculated fractions of proteins
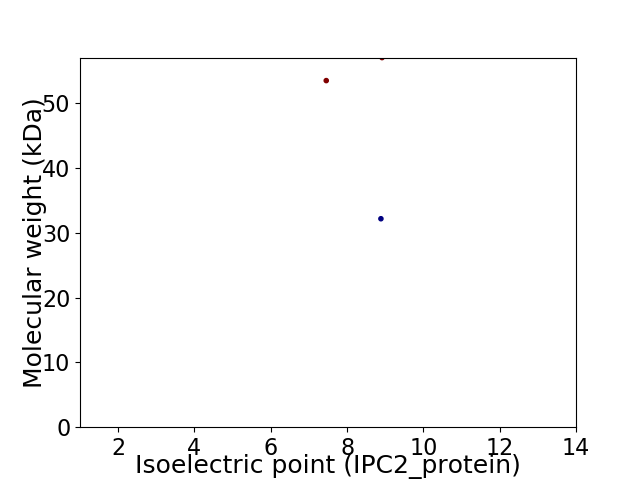
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KFX9|A0A1L3KFX9_9VIRU C2H2-type domain-containing protein OS=Beihai tombus-like virus 9 OX=1922730 PE=4 SV=1
MM1 pKa = 7.46NNSQSTFSPVHH12 pKa = 5.88KK13 pKa = 10.45CPVCGKK19 pKa = 9.6VFSSKK24 pKa = 10.46RR25 pKa = 11.84GLAQHH30 pKa = 6.49GQAKK34 pKa = 10.21HH35 pKa = 5.67GWTKK39 pKa = 8.41PQRR42 pKa = 11.84RR43 pKa = 11.84NRR45 pKa = 11.84SNDD48 pKa = 2.84QPTGGKK54 pKa = 10.29GLVPRR59 pKa = 11.84GNPGNTMGNKK69 pKa = 9.37KK70 pKa = 10.19RR71 pKa = 11.84VDD73 pKa = 3.84QIDD76 pKa = 5.55LITTPNWGYY85 pKa = 9.99CEE87 pKa = 4.51KK88 pKa = 10.69VFKK91 pKa = 10.86KK92 pKa = 10.4LISVGCTLDD101 pKa = 3.46GAAHH105 pKa = 5.84VVHH108 pKa = 6.92AVDD111 pKa = 4.07PACPEE116 pKa = 4.61GIPWNGIPDD125 pKa = 3.82EE126 pKa = 4.29NTTRR130 pKa = 11.84ILSRR134 pKa = 11.84RR135 pKa = 11.84IISINTITKK144 pKa = 9.19AHH146 pKa = 5.96SAVGNWDD153 pKa = 3.8FALMTCWSPHH163 pKa = 6.28AEE165 pKa = 3.82FVWFQQKK172 pKa = 8.76TGGDD176 pKa = 3.26WSGATQGQHH185 pKa = 5.46WDD187 pKa = 3.65CKK189 pKa = 10.11LSRR192 pKa = 11.84FSRR195 pKa = 11.84LDD197 pKa = 3.23WQSVTRR203 pKa = 11.84FRR205 pKa = 11.84QAFGSTTIHH214 pKa = 6.39MDD216 pKa = 3.4ASATTTEE223 pKa = 4.1GTVYY227 pKa = 10.97SMQKK231 pKa = 9.72SPEE234 pKa = 3.93RR235 pKa = 11.84YY236 pKa = 9.54LEE238 pKa = 3.96NEE240 pKa = 4.28YY241 pKa = 11.2KK242 pKa = 10.57SGLVNKK248 pKa = 8.09TVNKK252 pKa = 9.87PIATVKK258 pKa = 10.15IEE260 pKa = 4.31HH261 pKa = 7.32DD262 pKa = 3.99SDD264 pKa = 5.79SDD266 pKa = 4.09DD267 pKa = 3.45SWISAPTTSTATKK280 pKa = 9.79SKK282 pKa = 10.31VVSPTGADD290 pKa = 2.92EE291 pKa = 4.08VTARR295 pKa = 11.84LLCEE299 pKa = 4.14YY300 pKa = 10.06CGKK303 pKa = 7.86WTQADD308 pKa = 4.07MAFMDD313 pKa = 4.13PGFYY317 pKa = 9.62TAEE320 pKa = 4.09AFKK323 pKa = 10.9GVYY326 pKa = 8.9SVCRR330 pKa = 11.84LAGPRR335 pKa = 11.84VWSVGSWQDD344 pKa = 3.01EE345 pKa = 4.36GGVDD349 pKa = 3.32RR350 pKa = 11.84WRR352 pKa = 11.84VVAPLGTHH360 pKa = 6.8HH361 pKa = 7.57DD362 pKa = 3.91FAKK365 pKa = 10.62SKK367 pKa = 11.04SPLVSNDD374 pKa = 2.04IFQPFTNGWVIFTNLDD390 pKa = 3.25PKK392 pKa = 10.7AQLVIKK398 pKa = 7.18TVRR401 pKa = 11.84GYY403 pKa = 10.77EE404 pKa = 3.81MEE406 pKa = 4.31VNSEE410 pKa = 4.25SIAALDD416 pKa = 3.53THH418 pKa = 6.88FSPDD422 pKa = 3.59SDD424 pKa = 3.56PLALHH429 pKa = 7.04FEE431 pKa = 4.43TQLLRR436 pKa = 11.84GLQSGGPASDD446 pKa = 4.09NDD448 pKa = 3.72FGDD451 pKa = 3.89ILSGALDD458 pKa = 3.61VLLDD462 pKa = 3.75GLTATGGPVGAVLGPAGKK480 pKa = 9.94LLKK483 pKa = 9.59KK484 pKa = 8.71TFWRR488 pKa = 4.11
MM1 pKa = 7.46NNSQSTFSPVHH12 pKa = 5.88KK13 pKa = 10.45CPVCGKK19 pKa = 9.6VFSSKK24 pKa = 10.46RR25 pKa = 11.84GLAQHH30 pKa = 6.49GQAKK34 pKa = 10.21HH35 pKa = 5.67GWTKK39 pKa = 8.41PQRR42 pKa = 11.84RR43 pKa = 11.84NRR45 pKa = 11.84SNDD48 pKa = 2.84QPTGGKK54 pKa = 10.29GLVPRR59 pKa = 11.84GNPGNTMGNKK69 pKa = 9.37KK70 pKa = 10.19RR71 pKa = 11.84VDD73 pKa = 3.84QIDD76 pKa = 5.55LITTPNWGYY85 pKa = 9.99CEE87 pKa = 4.51KK88 pKa = 10.69VFKK91 pKa = 10.86KK92 pKa = 10.4LISVGCTLDD101 pKa = 3.46GAAHH105 pKa = 5.84VVHH108 pKa = 6.92AVDD111 pKa = 4.07PACPEE116 pKa = 4.61GIPWNGIPDD125 pKa = 3.82EE126 pKa = 4.29NTTRR130 pKa = 11.84ILSRR134 pKa = 11.84RR135 pKa = 11.84IISINTITKK144 pKa = 9.19AHH146 pKa = 5.96SAVGNWDD153 pKa = 3.8FALMTCWSPHH163 pKa = 6.28AEE165 pKa = 3.82FVWFQQKK172 pKa = 8.76TGGDD176 pKa = 3.26WSGATQGQHH185 pKa = 5.46WDD187 pKa = 3.65CKK189 pKa = 10.11LSRR192 pKa = 11.84FSRR195 pKa = 11.84LDD197 pKa = 3.23WQSVTRR203 pKa = 11.84FRR205 pKa = 11.84QAFGSTTIHH214 pKa = 6.39MDD216 pKa = 3.4ASATTTEE223 pKa = 4.1GTVYY227 pKa = 10.97SMQKK231 pKa = 9.72SPEE234 pKa = 3.93RR235 pKa = 11.84YY236 pKa = 9.54LEE238 pKa = 3.96NEE240 pKa = 4.28YY241 pKa = 11.2KK242 pKa = 10.57SGLVNKK248 pKa = 8.09TVNKK252 pKa = 9.87PIATVKK258 pKa = 10.15IEE260 pKa = 4.31HH261 pKa = 7.32DD262 pKa = 3.99SDD264 pKa = 5.79SDD266 pKa = 4.09DD267 pKa = 3.45SWISAPTTSTATKK280 pKa = 9.79SKK282 pKa = 10.31VVSPTGADD290 pKa = 2.92EE291 pKa = 4.08VTARR295 pKa = 11.84LLCEE299 pKa = 4.14YY300 pKa = 10.06CGKK303 pKa = 7.86WTQADD308 pKa = 4.07MAFMDD313 pKa = 4.13PGFYY317 pKa = 9.62TAEE320 pKa = 4.09AFKK323 pKa = 10.9GVYY326 pKa = 8.9SVCRR330 pKa = 11.84LAGPRR335 pKa = 11.84VWSVGSWQDD344 pKa = 3.01EE345 pKa = 4.36GGVDD349 pKa = 3.32RR350 pKa = 11.84WRR352 pKa = 11.84VVAPLGTHH360 pKa = 6.8HH361 pKa = 7.57DD362 pKa = 3.91FAKK365 pKa = 10.62SKK367 pKa = 11.04SPLVSNDD374 pKa = 2.04IFQPFTNGWVIFTNLDD390 pKa = 3.25PKK392 pKa = 10.7AQLVIKK398 pKa = 7.18TVRR401 pKa = 11.84GYY403 pKa = 10.77EE404 pKa = 3.81MEE406 pKa = 4.31VNSEE410 pKa = 4.25SIAALDD416 pKa = 3.53THH418 pKa = 6.88FSPDD422 pKa = 3.59SDD424 pKa = 3.56PLALHH429 pKa = 7.04FEE431 pKa = 4.43TQLLRR436 pKa = 11.84GLQSGGPASDD446 pKa = 4.09NDD448 pKa = 3.72FGDD451 pKa = 3.89ILSGALDD458 pKa = 3.61VLLDD462 pKa = 3.75GLTATGGPVGAVLGPAGKK480 pKa = 9.94LLKK483 pKa = 9.59KK484 pKa = 8.71TFWRR488 pKa = 4.11
Molecular weight: 53.45 kDa
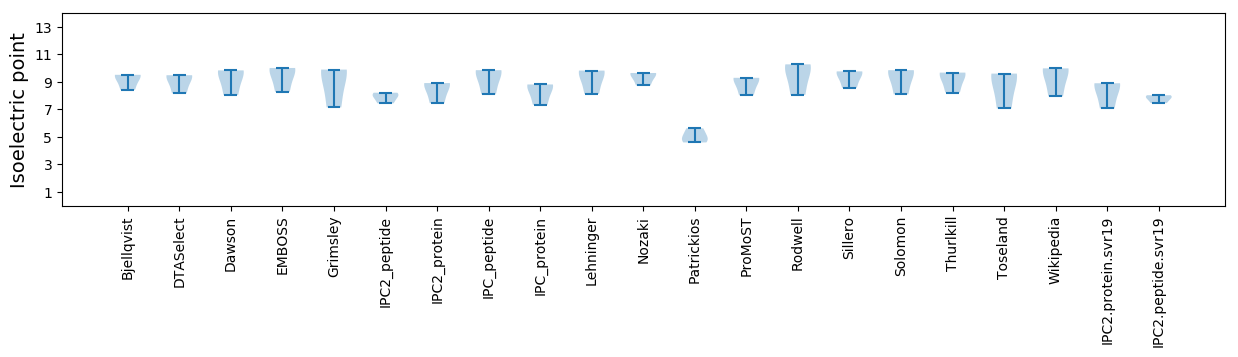
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KFX9|A0A1L3KFX9_9VIRU C2H2-type domain-containing protein OS=Beihai tombus-like virus 9 OX=1922730 PE=4 SV=1
MM1 pKa = 6.73VWDD4 pKa = 3.69YY5 pKa = 11.61AINQLDD11 pKa = 3.51MFYY14 pKa = 11.11YY15 pKa = 10.36NDD17 pKa = 4.0SGNQHH22 pKa = 5.11TVGVQLNNQHH32 pKa = 6.06KK33 pKa = 7.67TSSMIIHH40 pKa = 7.25NIPKK44 pKa = 10.03HH45 pKa = 4.61EE46 pKa = 4.28THH48 pKa = 6.74FSLAGIDD55 pKa = 5.71KK56 pKa = 9.84KK57 pKa = 11.22FCWFGRR63 pKa = 11.84KK64 pKa = 8.69IPLRR68 pKa = 11.84LPVRR72 pKa = 11.84LIYY75 pKa = 10.57SDD77 pKa = 4.17GSFTKK82 pKa = 10.29TKK84 pKa = 9.94FKK86 pKa = 10.74KK87 pKa = 10.45INTKK91 pKa = 10.42KK92 pKa = 10.34YY93 pKa = 10.5DD94 pKa = 4.04FIVNYY99 pKa = 10.08QGGSEE104 pKa = 4.23YY105 pKa = 11.0EE106 pKa = 4.19HH107 pKa = 6.64VLPQPEE113 pKa = 4.23PQVPIEE119 pKa = 3.74PHH121 pKa = 5.96HH122 pKa = 6.61ANKK125 pKa = 10.4EE126 pKa = 4.27PTSHH130 pKa = 7.12KK131 pKa = 10.6EE132 pKa = 3.58VDD134 pKa = 3.85EE135 pKa = 4.38TGMAHH140 pKa = 6.13MDD142 pKa = 3.24KK143 pKa = 10.45TKK145 pKa = 10.7TMLNHH150 pKa = 6.08TQPKK154 pKa = 9.64LKK156 pKa = 10.02VRR158 pKa = 11.84QRR160 pKa = 11.84ARR162 pKa = 11.84TYY164 pKa = 10.56FEE166 pKa = 3.88HH167 pKa = 6.6AQAITEE173 pKa = 4.31EE174 pKa = 3.93IRR176 pKa = 11.84LNICNVQGYY185 pKa = 8.26PCDD188 pKa = 3.17ATYY191 pKa = 11.07QLGVAKK197 pKa = 10.39GIEE200 pKa = 3.91IAKK203 pKa = 9.67KK204 pKa = 10.0YY205 pKa = 8.55NWQRR209 pKa = 11.84FTQSEE214 pKa = 4.1RR215 pKa = 11.84TEE217 pKa = 4.35IIRR220 pKa = 11.84KK221 pKa = 8.58SAAAANVPDD230 pKa = 4.2ATDD233 pKa = 3.44DD234 pKa = 3.62QIRR237 pKa = 11.84NMFKK241 pKa = 9.12ITAKK245 pKa = 9.73WIKK248 pKa = 9.41KK249 pKa = 7.95QNEE252 pKa = 3.93FTTTGNAGNLNGINGKK268 pKa = 9.03VAVSLPTKK276 pKa = 10.29KK277 pKa = 10.29AGKK280 pKa = 9.75AA281 pKa = 3.24
MM1 pKa = 6.73VWDD4 pKa = 3.69YY5 pKa = 11.61AINQLDD11 pKa = 3.51MFYY14 pKa = 11.11YY15 pKa = 10.36NDD17 pKa = 4.0SGNQHH22 pKa = 5.11TVGVQLNNQHH32 pKa = 6.06KK33 pKa = 7.67TSSMIIHH40 pKa = 7.25NIPKK44 pKa = 10.03HH45 pKa = 4.61EE46 pKa = 4.28THH48 pKa = 6.74FSLAGIDD55 pKa = 5.71KK56 pKa = 9.84KK57 pKa = 11.22FCWFGRR63 pKa = 11.84KK64 pKa = 8.69IPLRR68 pKa = 11.84LPVRR72 pKa = 11.84LIYY75 pKa = 10.57SDD77 pKa = 4.17GSFTKK82 pKa = 10.29TKK84 pKa = 9.94FKK86 pKa = 10.74KK87 pKa = 10.45INTKK91 pKa = 10.42KK92 pKa = 10.34YY93 pKa = 10.5DD94 pKa = 4.04FIVNYY99 pKa = 10.08QGGSEE104 pKa = 4.23YY105 pKa = 11.0EE106 pKa = 4.19HH107 pKa = 6.64VLPQPEE113 pKa = 4.23PQVPIEE119 pKa = 3.74PHH121 pKa = 5.96HH122 pKa = 6.61ANKK125 pKa = 10.4EE126 pKa = 4.27PTSHH130 pKa = 7.12KK131 pKa = 10.6EE132 pKa = 3.58VDD134 pKa = 3.85EE135 pKa = 4.38TGMAHH140 pKa = 6.13MDD142 pKa = 3.24KK143 pKa = 10.45TKK145 pKa = 10.7TMLNHH150 pKa = 6.08TQPKK154 pKa = 9.64LKK156 pKa = 10.02VRR158 pKa = 11.84QRR160 pKa = 11.84ARR162 pKa = 11.84TYY164 pKa = 10.56FEE166 pKa = 3.88HH167 pKa = 6.6AQAITEE173 pKa = 4.31EE174 pKa = 3.93IRR176 pKa = 11.84LNICNVQGYY185 pKa = 8.26PCDD188 pKa = 3.17ATYY191 pKa = 11.07QLGVAKK197 pKa = 10.39GIEE200 pKa = 3.91IAKK203 pKa = 9.67KK204 pKa = 10.0YY205 pKa = 8.55NWQRR209 pKa = 11.84FTQSEE214 pKa = 4.1RR215 pKa = 11.84TEE217 pKa = 4.35IIRR220 pKa = 11.84KK221 pKa = 8.58SAAAANVPDD230 pKa = 4.2ATDD233 pKa = 3.44DD234 pKa = 3.62QIRR237 pKa = 11.84NMFKK241 pKa = 9.12ITAKK245 pKa = 9.73WIKK248 pKa = 9.41KK249 pKa = 7.95QNEE252 pKa = 3.93FTTTGNAGNLNGINGKK268 pKa = 9.03VAVSLPTKK276 pKa = 10.29KK277 pKa = 10.29AGKK280 pKa = 9.75AA281 pKa = 3.24
Molecular weight: 32.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1264 |
281 |
495 |
421.3 |
47.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.12 ± 0.025 | 1.741 ± 0.204 |
5.696 ± 0.432 | 4.193 ± 0.372 |
4.193 ± 0.126 | 7.516 ± 0.749 |
3.56 ± 0.301 | 5.538 ± 0.742 |
7.674 ± 0.839 | 6.883 ± 0.874 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.294 ± 0.292 | 4.984 ± 0.64 |
4.589 ± 0.469 | 3.877 ± 0.634 |
5.38 ± 0.812 | 5.854 ± 0.992 |
7.041 ± 0.742 | 6.171 ± 0.453 |
2.453 ± 0.38 | 3.244 ± 0.697 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
