
Rhizoctonia solani
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetes incertae sedis; Cantharellales; Ceratobasidiaceae; Rhizoctonia
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 11878 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
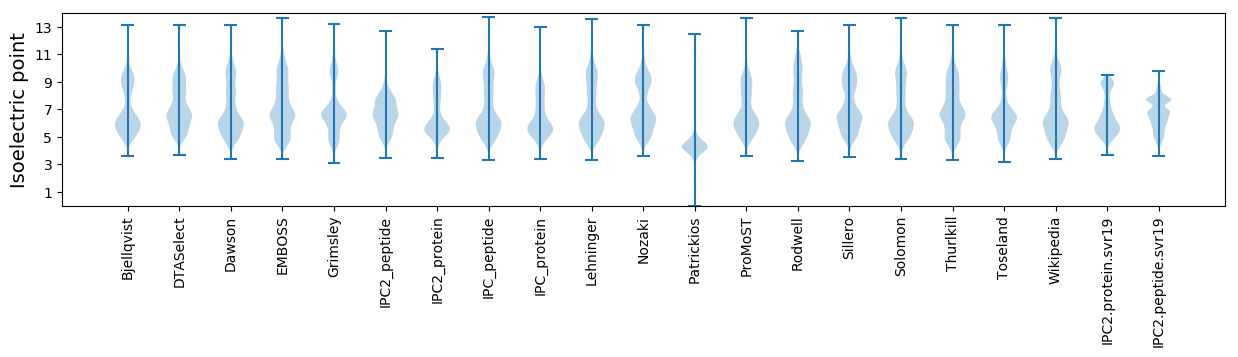
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A0K6FSL0|A0A0K6FSL0_9AGAM Retrotransposon-derived protein PEG10 OS=Rhizoctonia solani OX=456999 GN=RSOLAG22IIIB_08230 PE=4 SV=1
MM1 pKa = 7.58SSILAIALFAQVIAANPIYY20 pKa = 10.17RR21 pKa = 11.84RR22 pKa = 11.84QDD24 pKa = 2.94NGTTTITEE32 pKa = 4.12VTEE35 pKa = 4.42TPTATSTLSANATSITTDD53 pKa = 4.56PITFTVPTDD62 pKa = 3.35TTAIPITFSTAITSVAPSPTEE83 pKa = 3.99GSDD86 pKa = 3.75DD87 pKa = 3.46GSYY90 pKa = 9.48WYY92 pKa = 10.61CSFPGSEE99 pKa = 4.11ATATATATATAITTDD114 pKa = 3.47PFSTYY119 pKa = 9.48TSIDD123 pKa = 3.73DD124 pKa = 3.77TTVIADD130 pKa = 3.54PTFTDD135 pKa = 3.29SATEE139 pKa = 3.77ILTPTSTLTVPDD151 pKa = 3.58VTEE154 pKa = 4.15TATGTLSDD162 pKa = 4.33TEE164 pKa = 4.81LPTATEE170 pKa = 4.21TATDD174 pKa = 3.75TDD176 pKa = 3.79VTTATEE182 pKa = 4.35STTDD186 pKa = 3.12TDD188 pKa = 4.72IITATITAVTPTATDD203 pKa = 3.25TAIITDD209 pKa = 3.68VTSVSEE215 pKa = 4.11TDD217 pKa = 3.32TFIDD221 pKa = 3.65TFTDD225 pKa = 3.37TFTDD229 pKa = 3.59TFTDD233 pKa = 3.01TSTFVEE239 pKa = 4.81ATATTGPIVTITSVPSRR256 pKa = 11.84VQTYY260 pKa = 8.37TIVGPSTTAVIKK272 pKa = 9.85CIRR275 pKa = 11.84IRR277 pKa = 11.84GGPGRR282 pKa = 11.84PHH284 pKa = 6.56SPYY287 pKa = 9.67PHH289 pKa = 6.8PHH291 pKa = 7.49PYY293 pKa = 9.49PHH295 pKa = 7.56PNPPHH300 pKa = 6.6HH301 pKa = 7.21SSGSPQTFSGFPTATTDD318 pKa = 3.6VLGPTATEE326 pKa = 4.02TLPATSIVSFYY337 pKa = 10.45TAVPTTTFGVVGG349 pKa = 3.93
MM1 pKa = 7.58SSILAIALFAQVIAANPIYY20 pKa = 10.17RR21 pKa = 11.84RR22 pKa = 11.84QDD24 pKa = 2.94NGTTTITEE32 pKa = 4.12VTEE35 pKa = 4.42TPTATSTLSANATSITTDD53 pKa = 4.56PITFTVPTDD62 pKa = 3.35TTAIPITFSTAITSVAPSPTEE83 pKa = 3.99GSDD86 pKa = 3.75DD87 pKa = 3.46GSYY90 pKa = 9.48WYY92 pKa = 10.61CSFPGSEE99 pKa = 4.11ATATATATATAITTDD114 pKa = 3.47PFSTYY119 pKa = 9.48TSIDD123 pKa = 3.73DD124 pKa = 3.77TTVIADD130 pKa = 3.54PTFTDD135 pKa = 3.29SATEE139 pKa = 3.77ILTPTSTLTVPDD151 pKa = 3.58VTEE154 pKa = 4.15TATGTLSDD162 pKa = 4.33TEE164 pKa = 4.81LPTATEE170 pKa = 4.21TATDD174 pKa = 3.75TDD176 pKa = 3.79VTTATEE182 pKa = 4.35STTDD186 pKa = 3.12TDD188 pKa = 4.72IITATITAVTPTATDD203 pKa = 3.25TAIITDD209 pKa = 3.68VTSVSEE215 pKa = 4.11TDD217 pKa = 3.32TFIDD221 pKa = 3.65TFTDD225 pKa = 3.37TFTDD229 pKa = 3.59TFTDD233 pKa = 3.01TSTFVEE239 pKa = 4.81ATATTGPIVTITSVPSRR256 pKa = 11.84VQTYY260 pKa = 8.37TIVGPSTTAVIKK272 pKa = 9.85CIRR275 pKa = 11.84IRR277 pKa = 11.84GGPGRR282 pKa = 11.84PHH284 pKa = 6.56SPYY287 pKa = 9.67PHH289 pKa = 6.8PHH291 pKa = 7.49PYY293 pKa = 9.49PHH295 pKa = 7.56PNPPHH300 pKa = 6.6HH301 pKa = 7.21SSGSPQTFSGFPTATTDD318 pKa = 3.6VLGPTATEE326 pKa = 4.02TLPATSIVSFYY337 pKa = 10.45TAVPTTTFGVVGG349 pKa = 3.93
Molecular weight: 36.16 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K6FYK2|A0A0K6FYK2_9AGAM RRM domain-containing protein OS=Rhizoctonia solani OX=456999 GN=RSOLAG22IIIB_04502 PE=4 SV=1
MM1 pKa = 7.04QPKK4 pKa = 9.76LLSVLITFFGLATASPIMQGRR25 pKa = 11.84AVPSAKK31 pKa = 9.71PHH33 pKa = 6.36PPPSVTASLPPHH45 pKa = 6.87PSLSAFPSFSPLPPPHH61 pKa = 6.77HH62 pKa = 6.73SFVGRR67 pKa = 11.84QAPSSFPPPPPPPSSLPPRR86 pKa = 11.84PTFSPRR92 pKa = 11.84PPPPPPSLAGRR103 pKa = 11.84QAPSSLRR110 pKa = 11.84PPPPPPSSLPPRR122 pKa = 11.84PTFSPRR128 pKa = 11.84PPPPAPLAGRR138 pKa = 11.84QAPSSFPHH146 pKa = 6.77PPPPPSSLPPRR157 pKa = 11.84PTFSPLPPPPPPSLAARR174 pKa = 11.84QAPSSFPPPPPPPSSLPPRR193 pKa = 11.84PTFSPRR199 pKa = 11.84PPPPPPSLAGRR210 pKa = 11.84QAPSSRR216 pKa = 11.84PPPPPPSSLPPRR228 pKa = 11.84PTLSLPPPPPSPTASAA244 pKa = 3.77
MM1 pKa = 7.04QPKK4 pKa = 9.76LLSVLITFFGLATASPIMQGRR25 pKa = 11.84AVPSAKK31 pKa = 9.71PHH33 pKa = 6.36PPPSVTASLPPHH45 pKa = 6.87PSLSAFPSFSPLPPPHH61 pKa = 6.77HH62 pKa = 6.73SFVGRR67 pKa = 11.84QAPSSFPPPPPPPSSLPPRR86 pKa = 11.84PTFSPRR92 pKa = 11.84PPPPPPSLAGRR103 pKa = 11.84QAPSSLRR110 pKa = 11.84PPPPPPSSLPPRR122 pKa = 11.84PTFSPRR128 pKa = 11.84PPPPAPLAGRR138 pKa = 11.84QAPSSFPHH146 pKa = 6.77PPPPPSSLPPRR157 pKa = 11.84PTFSPLPPPPPPSLAARR174 pKa = 11.84QAPSSFPPPPPPPSSLPPRR193 pKa = 11.84PTFSPRR199 pKa = 11.84PPPPPPSLAGRR210 pKa = 11.84QAPSSRR216 pKa = 11.84PPPPPPSSLPPRR228 pKa = 11.84PTLSLPPPPPSPTASAA244 pKa = 3.77
Molecular weight: 25.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6275154 |
66 |
5018 |
528.3 |
57.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.418 ± 0.023 | 1.183 ± 0.008 |
5.396 ± 0.016 | 5.79 ± 0.021 |
3.432 ± 0.011 | 6.67 ± 0.022 |
2.448 ± 0.01 | 4.938 ± 0.014 |
4.458 ± 0.019 | 8.801 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.007 ± 0.008 | 3.602 ± 0.012 |
6.528 ± 0.034 | 3.749 ± 0.014 |
5.996 ± 0.02 | 8.387 ± 0.025 |
6.145 ± 0.017 | 6.17 ± 0.017 |
1.45 ± 0.008 | 2.709 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
