
Byssochlamys spectabilis (Paecilomyces variotii)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta;
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
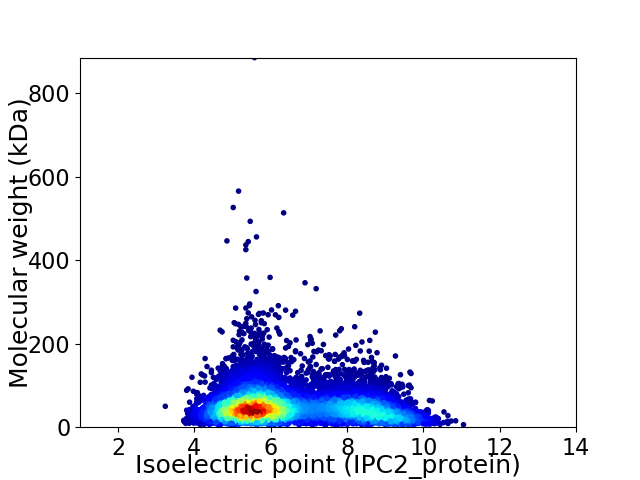
Virtual 2D-PAGE plot for 9264 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
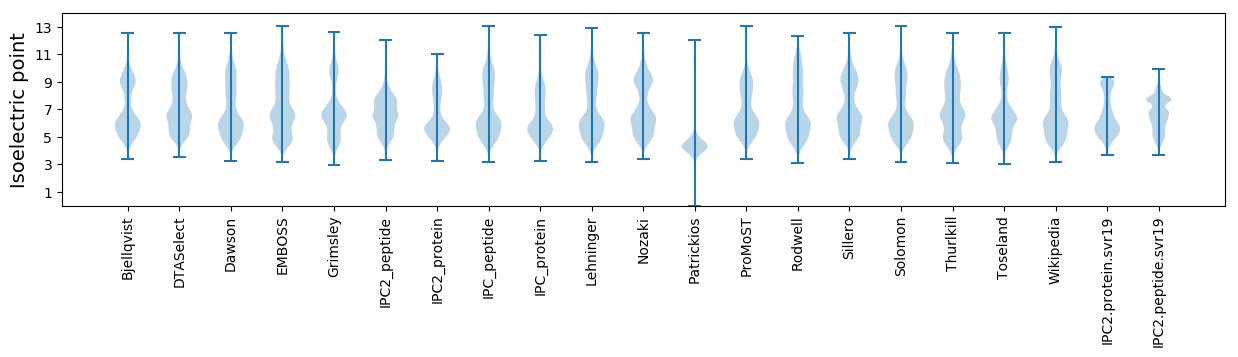
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A443HWV5|A0A443HWV5_BYSSP Dipeptidyl-peptidase IV OS=Byssochlamys spectabilis OX=264951 GN=C8Q69DRAFT_464790 PE=3 SV=1
GG1 pKa = 6.89QGYY4 pKa = 7.82NQPRR8 pKa = 11.84LSQICQEE15 pKa = 4.13TSLDD19 pKa = 3.61IINIGFVNVFPDD31 pKa = 3.76VGQGGWPGSNFGNQCNGQTYY51 pKa = 9.59TNADD55 pKa = 3.51GEE57 pKa = 4.56ATQLLSGCQQIADD70 pKa = 5.4DD71 pKa = 5.56IPVCQQAGKK80 pKa = 10.42KK81 pKa = 9.43VFISLGGAYY90 pKa = 9.13PADD93 pKa = 3.67GQIIDD98 pKa = 4.47NDD100 pKa = 3.42DD101 pKa = 3.53SARR104 pKa = 11.84DD105 pKa = 3.49FADD108 pKa = 3.97FLWGAFGPEE117 pKa = 3.22TDD119 pKa = 3.61AWVAQNGPRR128 pKa = 11.84PFGDD132 pKa = 3.11AVVDD136 pKa = 4.34GFDD139 pKa = 5.15FDD141 pKa = 4.57IEE143 pKa = 4.81HH144 pKa = 6.88NGPTGYY150 pKa = 10.77AALARR155 pKa = 11.84RR156 pKa = 11.84LRR158 pKa = 11.84DD159 pKa = 3.46NFSEE163 pKa = 4.79FPSKK167 pKa = 10.61QFYY170 pKa = 10.82LSAAPQCLSPDD181 pKa = 3.54AQLSNAIQFASFDD194 pKa = 4.54FIWVQFYY201 pKa = 8.58NTDD204 pKa = 3.59PCSARR209 pKa = 11.84AWVDD213 pKa = 2.96GDD215 pKa = 3.73VTSDD219 pKa = 3.32FTFDD223 pKa = 2.65AWIAAIKK230 pKa = 10.66LGGNPSAKK238 pKa = 10.28LFIGLPADD246 pKa = 3.53PTQAYY251 pKa = 8.85YY252 pKa = 10.93VSPDD256 pKa = 3.24EE257 pKa = 4.67VEE259 pKa = 4.41TLVEE263 pKa = 4.55EE264 pKa = 4.33YY265 pKa = 9.62MGRR268 pKa = 11.84YY269 pKa = 8.44PEE271 pKa = 4.15NFGGIMIWEE280 pKa = 4.19ATASDD285 pKa = 4.05EE286 pKa = 4.12NQIDD290 pKa = 3.79GRR292 pKa = 11.84SYY294 pKa = 9.32AANMKK299 pKa = 10.28SVLLL303 pKa = 4.11
GG1 pKa = 6.89QGYY4 pKa = 7.82NQPRR8 pKa = 11.84LSQICQEE15 pKa = 4.13TSLDD19 pKa = 3.61IINIGFVNVFPDD31 pKa = 3.76VGQGGWPGSNFGNQCNGQTYY51 pKa = 9.59TNADD55 pKa = 3.51GEE57 pKa = 4.56ATQLLSGCQQIADD70 pKa = 5.4DD71 pKa = 5.56IPVCQQAGKK80 pKa = 10.42KK81 pKa = 9.43VFISLGGAYY90 pKa = 9.13PADD93 pKa = 3.67GQIIDD98 pKa = 4.47NDD100 pKa = 3.42DD101 pKa = 3.53SARR104 pKa = 11.84DD105 pKa = 3.49FADD108 pKa = 3.97FLWGAFGPEE117 pKa = 3.22TDD119 pKa = 3.61AWVAQNGPRR128 pKa = 11.84PFGDD132 pKa = 3.11AVVDD136 pKa = 4.34GFDD139 pKa = 5.15FDD141 pKa = 4.57IEE143 pKa = 4.81HH144 pKa = 6.88NGPTGYY150 pKa = 10.77AALARR155 pKa = 11.84RR156 pKa = 11.84LRR158 pKa = 11.84DD159 pKa = 3.46NFSEE163 pKa = 4.79FPSKK167 pKa = 10.61QFYY170 pKa = 10.82LSAAPQCLSPDD181 pKa = 3.54AQLSNAIQFASFDD194 pKa = 4.54FIWVQFYY201 pKa = 8.58NTDD204 pKa = 3.59PCSARR209 pKa = 11.84AWVDD213 pKa = 2.96GDD215 pKa = 3.73VTSDD219 pKa = 3.32FTFDD223 pKa = 2.65AWIAAIKK230 pKa = 10.66LGGNPSAKK238 pKa = 10.28LFIGLPADD246 pKa = 3.53PTQAYY251 pKa = 8.85YY252 pKa = 10.93VSPDD256 pKa = 3.24EE257 pKa = 4.67VEE259 pKa = 4.41TLVEE263 pKa = 4.55EE264 pKa = 4.33YY265 pKa = 9.62MGRR268 pKa = 11.84YY269 pKa = 8.44PEE271 pKa = 4.15NFGGIMIWEE280 pKa = 4.19ATASDD285 pKa = 4.05EE286 pKa = 4.12NQIDD290 pKa = 3.79GRR292 pKa = 11.84SYY294 pKa = 9.32AANMKK299 pKa = 10.28SVLLL303 pKa = 4.11
Molecular weight: 32.98 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A443HL20|A0A443HL20_BYSSP Tyrosine--tRNA ligase OS=Byssochlamys spectabilis OX=264951 GN=C8Q69DRAFT_479466 PE=3 SV=1
SS1 pKa = 6.85HH2 pKa = 7.5KK3 pKa = 10.71SFRR6 pKa = 11.84TKK8 pKa = 10.45QKK10 pKa = 9.84LAKK13 pKa = 9.55AQKK16 pKa = 8.59QNRR19 pKa = 11.84PIPQWIRR26 pKa = 11.84LRR28 pKa = 11.84TGNTIRR34 pKa = 11.84YY35 pKa = 5.79NAKK38 pKa = 8.89RR39 pKa = 11.84RR40 pKa = 11.84HH41 pKa = 4.14WRR43 pKa = 11.84KK44 pKa = 7.41TRR46 pKa = 11.84LGII49 pKa = 4.46
SS1 pKa = 6.85HH2 pKa = 7.5KK3 pKa = 10.71SFRR6 pKa = 11.84TKK8 pKa = 10.45QKK10 pKa = 9.84LAKK13 pKa = 9.55AQKK16 pKa = 8.59QNRR19 pKa = 11.84PIPQWIRR26 pKa = 11.84LRR28 pKa = 11.84TGNTIRR34 pKa = 11.84YY35 pKa = 5.79NAKK38 pKa = 8.89RR39 pKa = 11.84RR40 pKa = 11.84HH41 pKa = 4.14WRR43 pKa = 11.84KK44 pKa = 7.41TRR46 pKa = 11.84LGII49 pKa = 4.46
Molecular weight: 6.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4547040 |
49 |
7905 |
490.8 |
54.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.322 ± 0.019 | 1.182 ± 0.01 |
5.693 ± 0.017 | 6.402 ± 0.024 |
3.714 ± 0.017 | 6.75 ± 0.02 |
2.333 ± 0.01 | 5.037 ± 0.017 |
4.883 ± 0.023 | 8.879 ± 0.023 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.072 ± 0.009 | 3.645 ± 0.012 |
6.09 ± 0.028 | 3.974 ± 0.018 |
6.365 ± 0.022 | 8.514 ± 0.034 |
5.824 ± 0.016 | 6.064 ± 0.017 |
1.414 ± 0.009 | 2.842 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
