
Ochrobactrum intermedium 229E
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Brucellaceae; Brucella/Ochrobactrum group; Brucella; Brucella intermedia
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
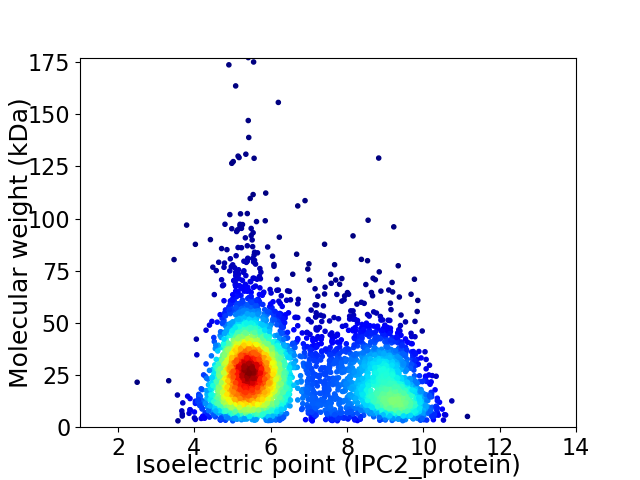
Virtual 2D-PAGE plot for 4249 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|U4V7R8|U4V7R8_9RHIZ Dihydroxy-acid dehydratase OS=Ochrobactrum intermedium 229E OX=1337887 GN=Q644_25235 PE=3 SV=1
SS1 pKa = 7.36DD2 pKa = 3.89AATTTVNVLVDD13 pKa = 4.24GVPYY17 pKa = 10.53AAVSNGDD24 pKa = 3.82GTWTASVPGSALAGAATPTVTAEE47 pKa = 4.19VVFADD52 pKa = 4.68AAGNDD57 pKa = 3.89AAPVTSTQTYY67 pKa = 7.88TVDD70 pKa = 3.28VTPPTTDD77 pKa = 2.97VAITDD82 pKa = 4.04PEE84 pKa = 4.95GDD86 pKa = 4.16NIPTASGTTEE96 pKa = 4.06PGSTVVVTWPDD107 pKa = 3.14GTTSQVTADD116 pKa = 3.74GTTGAWTATAVLAQPAGTVSVAVTDD141 pKa = 3.94PAGNPGSGSGGWTPSTTTPGAADD164 pKa = 3.95DD165 pKa = 4.3LAAASVDD172 pKa = 3.76VVPTEE177 pKa = 4.07TAVDD181 pKa = 3.73NGDD184 pKa = 3.1ATYY187 pKa = 11.41LLGIGIGLGIIDD199 pKa = 4.68LQATVLGVPSVGFTITEE216 pKa = 4.08GHH218 pKa = 5.52TQDD221 pKa = 3.28LTFAFGGLANVGVLGDD237 pKa = 3.83YY238 pKa = 10.46QVVVQKK244 pKa = 10.2WDD246 pKa = 3.3ALTGQWTSVDD256 pKa = 3.36GSATGGSILNIGLLDD271 pKa = 3.97GGANGVVIPDD281 pKa = 3.67MDD283 pKa = 4.29AGQYY287 pKa = 9.26RR288 pKa = 11.84AFMVYY293 pKa = 10.29NGVGLSVLTTMTVSGLDD310 pKa = 3.0HH311 pKa = 7.63DD312 pKa = 5.48YY313 pKa = 11.24INTSIVTGEE322 pKa = 3.84ATGNVLDD329 pKa = 4.49NDD331 pKa = 4.19GGAAAAGHH339 pKa = 6.48IVTSVNFNGTDD350 pKa = 3.54YY351 pKa = 11.0PVAAGPAGTTIIGQYY366 pKa = 8.35GQLVIHH372 pKa = 6.45QDD374 pKa = 2.76GSYY377 pKa = 9.95TYY379 pKa = 9.99TPNADD384 pKa = 2.75GTAIGKK390 pKa = 9.2VDD392 pKa = 3.34QFTYY396 pKa = 10.15TLHH399 pKa = 7.55DD400 pKa = 3.7PVANTNTQAILYY412 pKa = 9.18VRR414 pKa = 11.84IDD416 pKa = 3.21SDD418 pKa = 4.12GQGGLVWNDD427 pKa = 2.86ADD429 pKa = 4.25LGADD433 pKa = 3.21ATYY436 pKa = 11.1SVAATNDD443 pKa = 2.99ATEE446 pKa = 5.42IDD448 pKa = 3.92ITWINPTDD456 pKa = 3.26TAYY459 pKa = 10.77FDD461 pKa = 3.5QAQPLIGGALGTVTANSNQFTIGSNMDD488 pKa = 3.03ATGSVVVSVTAAALASGTVTIQKK511 pKa = 10.18LVGSVWQDD519 pKa = 3.11VNTDD523 pKa = 3.01NAFNVAVGVLGNVKK537 pKa = 9.72TIDD540 pKa = 3.55IGSLDD545 pKa = 4.5LDD547 pKa = 3.85AGTYY551 pKa = 9.28RR552 pKa = 11.84VHH554 pKa = 5.79TTLSGALAAISITTDD569 pKa = 3.08VNVTHH574 pKa = 7.47LDD576 pKa = 3.25QHH578 pKa = 6.89VISGDD583 pKa = 3.57PSISGSLVANDD594 pKa = 3.72GVVPEE599 pKa = 4.44GSRR602 pKa = 11.84VVVSDD607 pKa = 3.4GGTFVDD613 pKa = 5.34ASAAGTVIHH622 pKa = 6.66GVHH625 pKa = 6.88GDD627 pKa = 3.31LTVYY631 pKa = 11.23SNGTYY636 pKa = 9.87KK637 pKa = 10.75YY638 pKa = 10.47EE639 pKa = 4.35PLDD642 pKa = 3.44TLAYY646 pKa = 10.03ADD648 pKa = 4.86RR649 pKa = 11.84EE650 pKa = 4.57GTDD653 pKa = 2.86SFTYY657 pKa = 10.27KK658 pKa = 9.98IVLPGGYY665 pKa = 7.54EE666 pKa = 3.87TTAEE670 pKa = 4.31LVVQLHH676 pKa = 7.13DD677 pKa = 3.7GAAVTFTTEE686 pKa = 3.46TADD689 pKa = 3.64PVSAEE694 pKa = 4.25SFSADD699 pKa = 3.0TSGDD703 pKa = 3.56VVALGGLAGSDD714 pKa = 3.76ASDD717 pKa = 3.73SSPEE721 pKa = 3.91PAQHH725 pKa = 6.31NLSEE729 pKa = 4.86ALLLDD734 pKa = 4.41DD735 pKa = 5.08GGGEE739 pKa = 4.05IVLPSSDD746 pKa = 3.93SEE748 pKa = 4.53TGGGSSADD756 pKa = 3.4AAVSSTDD763 pKa = 2.98TDD765 pKa = 5.18LITVDD770 pKa = 3.81EE771 pKa = 5.05GSTVIDD777 pKa = 3.64PLAYY781 pKa = 8.42LTPDD785 pKa = 3.96PLRR788 pKa = 11.84QEE790 pKa = 4.9DD791 pKa = 4.57PPPHH795 pKa = 6.25SAHH798 pKa = 5.81MVV800 pKa = 3.15
SS1 pKa = 7.36DD2 pKa = 3.89AATTTVNVLVDD13 pKa = 4.24GVPYY17 pKa = 10.53AAVSNGDD24 pKa = 3.82GTWTASVPGSALAGAATPTVTAEE47 pKa = 4.19VVFADD52 pKa = 4.68AAGNDD57 pKa = 3.89AAPVTSTQTYY67 pKa = 7.88TVDD70 pKa = 3.28VTPPTTDD77 pKa = 2.97VAITDD82 pKa = 4.04PEE84 pKa = 4.95GDD86 pKa = 4.16NIPTASGTTEE96 pKa = 4.06PGSTVVVTWPDD107 pKa = 3.14GTTSQVTADD116 pKa = 3.74GTTGAWTATAVLAQPAGTVSVAVTDD141 pKa = 3.94PAGNPGSGSGGWTPSTTTPGAADD164 pKa = 3.95DD165 pKa = 4.3LAAASVDD172 pKa = 3.76VVPTEE177 pKa = 4.07TAVDD181 pKa = 3.73NGDD184 pKa = 3.1ATYY187 pKa = 11.41LLGIGIGLGIIDD199 pKa = 4.68LQATVLGVPSVGFTITEE216 pKa = 4.08GHH218 pKa = 5.52TQDD221 pKa = 3.28LTFAFGGLANVGVLGDD237 pKa = 3.83YY238 pKa = 10.46QVVVQKK244 pKa = 10.2WDD246 pKa = 3.3ALTGQWTSVDD256 pKa = 3.36GSATGGSILNIGLLDD271 pKa = 3.97GGANGVVIPDD281 pKa = 3.67MDD283 pKa = 4.29AGQYY287 pKa = 9.26RR288 pKa = 11.84AFMVYY293 pKa = 10.29NGVGLSVLTTMTVSGLDD310 pKa = 3.0HH311 pKa = 7.63DD312 pKa = 5.48YY313 pKa = 11.24INTSIVTGEE322 pKa = 3.84ATGNVLDD329 pKa = 4.49NDD331 pKa = 4.19GGAAAAGHH339 pKa = 6.48IVTSVNFNGTDD350 pKa = 3.54YY351 pKa = 11.0PVAAGPAGTTIIGQYY366 pKa = 8.35GQLVIHH372 pKa = 6.45QDD374 pKa = 2.76GSYY377 pKa = 9.95TYY379 pKa = 9.99TPNADD384 pKa = 2.75GTAIGKK390 pKa = 9.2VDD392 pKa = 3.34QFTYY396 pKa = 10.15TLHH399 pKa = 7.55DD400 pKa = 3.7PVANTNTQAILYY412 pKa = 9.18VRR414 pKa = 11.84IDD416 pKa = 3.21SDD418 pKa = 4.12GQGGLVWNDD427 pKa = 2.86ADD429 pKa = 4.25LGADD433 pKa = 3.21ATYY436 pKa = 11.1SVAATNDD443 pKa = 2.99ATEE446 pKa = 5.42IDD448 pKa = 3.92ITWINPTDD456 pKa = 3.26TAYY459 pKa = 10.77FDD461 pKa = 3.5QAQPLIGGALGTVTANSNQFTIGSNMDD488 pKa = 3.03ATGSVVVSVTAAALASGTVTIQKK511 pKa = 10.18LVGSVWQDD519 pKa = 3.11VNTDD523 pKa = 3.01NAFNVAVGVLGNVKK537 pKa = 9.72TIDD540 pKa = 3.55IGSLDD545 pKa = 4.5LDD547 pKa = 3.85AGTYY551 pKa = 9.28RR552 pKa = 11.84VHH554 pKa = 5.79TTLSGALAAISITTDD569 pKa = 3.08VNVTHH574 pKa = 7.47LDD576 pKa = 3.25QHH578 pKa = 6.89VISGDD583 pKa = 3.57PSISGSLVANDD594 pKa = 3.72GVVPEE599 pKa = 4.44GSRR602 pKa = 11.84VVVSDD607 pKa = 3.4GGTFVDD613 pKa = 5.34ASAAGTVIHH622 pKa = 6.66GVHH625 pKa = 6.88GDD627 pKa = 3.31LTVYY631 pKa = 11.23SNGTYY636 pKa = 9.87KK637 pKa = 10.75YY638 pKa = 10.47EE639 pKa = 4.35PLDD642 pKa = 3.44TLAYY646 pKa = 10.03ADD648 pKa = 4.86RR649 pKa = 11.84EE650 pKa = 4.57GTDD653 pKa = 2.86SFTYY657 pKa = 10.27KK658 pKa = 9.98IVLPGGYY665 pKa = 7.54EE666 pKa = 3.87TTAEE670 pKa = 4.31LVVQLHH676 pKa = 7.13DD677 pKa = 3.7GAAVTFTTEE686 pKa = 3.46TADD689 pKa = 3.64PVSAEE694 pKa = 4.25SFSADD699 pKa = 3.0TSGDD703 pKa = 3.56VVALGGLAGSDD714 pKa = 3.76ASDD717 pKa = 3.73SSPEE721 pKa = 3.91PAQHH725 pKa = 6.31NLSEE729 pKa = 4.86ALLLDD734 pKa = 4.41DD735 pKa = 5.08GGGEE739 pKa = 4.05IVLPSSDD746 pKa = 3.93SEE748 pKa = 4.53TGGGSSADD756 pKa = 3.4AAVSSTDD763 pKa = 2.98TDD765 pKa = 5.18LITVDD770 pKa = 3.81EE771 pKa = 5.05GSTVIDD777 pKa = 3.64PLAYY781 pKa = 8.42LTPDD785 pKa = 3.96PLRR788 pKa = 11.84QEE790 pKa = 4.9DD791 pKa = 4.57PPPHH795 pKa = 6.25SAHH798 pKa = 5.81MVV800 pKa = 3.15
Molecular weight: 80.34 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|U4V474|U4V474_9RHIZ Uncharacterized protein OS=Ochrobactrum intermedium 229E OX=1337887 GN=Q644_24875 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.09QPSKK9 pKa = 8.8IVRR12 pKa = 11.84KK13 pKa = 9.24RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.55GFRR19 pKa = 11.84ARR21 pKa = 11.84MATTGGRR28 pKa = 11.84KK29 pKa = 8.98VLAARR34 pKa = 11.84RR35 pKa = 11.84SRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.96RR41 pKa = 11.84LSAA44 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.09QPSKK9 pKa = 8.8IVRR12 pKa = 11.84KK13 pKa = 9.24RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.55GFRR19 pKa = 11.84ARR21 pKa = 11.84MATTGGRR28 pKa = 11.84KK29 pKa = 8.98VLAARR34 pKa = 11.84RR35 pKa = 11.84SRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.96RR41 pKa = 11.84LSAA44 pKa = 4.03
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1077194 |
29 |
1622 |
253.5 |
27.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.356 ± 0.045 | 0.812 ± 0.012 |
5.563 ± 0.032 | 5.847 ± 0.041 |
3.979 ± 0.028 | 8.367 ± 0.04 |
1.989 ± 0.018 | 5.849 ± 0.031 |
4.032 ± 0.032 | 9.851 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.618 ± 0.015 | 3.074 ± 0.022 |
4.943 ± 0.026 | 3.222 ± 0.02 |
6.577 ± 0.044 | 5.804 ± 0.028 |
5.224 ± 0.024 | 7.219 ± 0.033 |
1.278 ± 0.016 | 2.379 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
