
Circovirus-like genome DCCV-10
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 7.98
Get precalculated fractions of proteins
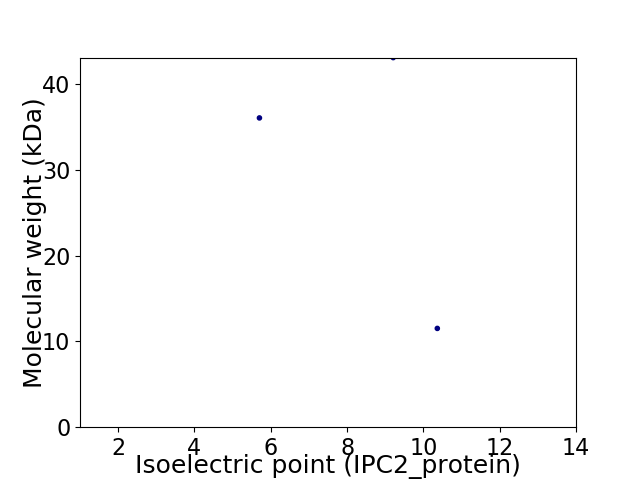
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
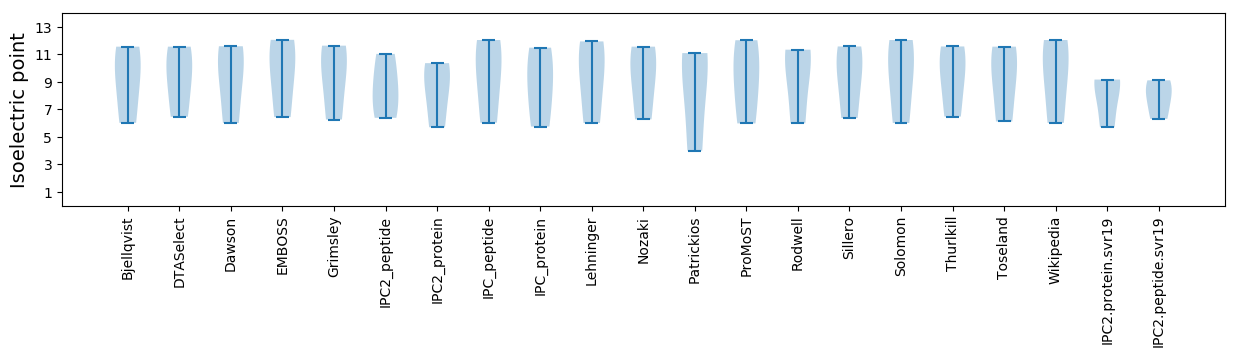
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A190WHE1|A0A190WHE1_9CIRC ATP-dependent helicase Rep OS=Circovirus-like genome DCCV-10 OX=1788438 PE=3 SV=1
MM1 pKa = 6.46TTRR4 pKa = 11.84IRR6 pKa = 11.84AWCFTLNNPTDD17 pKa = 3.66RR18 pKa = 11.84DD19 pKa = 3.54KK20 pKa = 11.43RR21 pKa = 11.84ALIALANTTRR31 pKa = 11.84YY32 pKa = 10.22VIAQLEE38 pKa = 4.3HH39 pKa = 6.95APTTGTPHH47 pKa = 5.39YY48 pKa = 8.8QGYY51 pKa = 7.88MVFHH55 pKa = 7.6DD56 pKa = 4.58GKK58 pKa = 11.0SFSALHH64 pKa = 6.09KK65 pKa = 10.59AMPRR69 pKa = 11.84AHH71 pKa = 6.94FEE73 pKa = 3.97PARR76 pKa = 11.84GTAQQNYY83 pKa = 9.89DD84 pKa = 4.04YY85 pKa = 9.02CTKK88 pKa = 10.63AADD91 pKa = 3.61EE92 pKa = 4.55GQPSNEE98 pKa = 3.81LVLEE102 pKa = 4.64HH103 pKa = 6.49GTLPQQGEE111 pKa = 4.36RR112 pKa = 11.84VDD114 pKa = 3.37ISQVRR119 pKa = 11.84EE120 pKa = 3.89LLRR123 pKa = 11.84SGVTYY128 pKa = 10.79RR129 pKa = 11.84EE130 pKa = 4.09LVDD133 pKa = 3.36MDD135 pKa = 3.9VNYY138 pKa = 10.35QVLRR142 pKa = 11.84IAQEE146 pKa = 3.74WLRR149 pKa = 11.84VHH151 pKa = 6.84EE152 pKa = 4.48PARR155 pKa = 11.84DD156 pKa = 3.85FKK158 pKa = 11.56PEE160 pKa = 3.69VRR162 pKa = 11.84WFYY165 pKa = 11.44GEE167 pKa = 4.04PGAGKK172 pKa = 8.06TRR174 pKa = 11.84AALEE178 pKa = 3.93WLEE181 pKa = 3.9SHH183 pKa = 6.66GEE185 pKa = 4.16VFTVEE190 pKa = 4.33TPAKK194 pKa = 9.98YY195 pKa = 8.51WDD197 pKa = 4.65GYY199 pKa = 10.94DD200 pKa = 3.29GHH202 pKa = 7.14NCVLFDD208 pKa = 5.79DD209 pKa = 5.51LRR211 pKa = 11.84ADD213 pKa = 3.09QCGFVRR219 pKa = 11.84MLRR222 pKa = 11.84LLDD225 pKa = 4.08RR226 pKa = 11.84YY227 pKa = 10.98AMRR230 pKa = 11.84VEE232 pKa = 4.22VKK234 pKa = 10.52GGSKK238 pKa = 8.18QLRR241 pKa = 11.84ARR243 pKa = 11.84YY244 pKa = 8.81IAITAPHH251 pKa = 6.83RR252 pKa = 11.84PEE254 pKa = 4.05EE255 pKa = 4.16IYY257 pKa = 9.43RR258 pKa = 11.84TSSEE262 pKa = 4.42NIVQLTRR269 pKa = 11.84RR270 pKa = 11.84ISSVTHH276 pKa = 4.73VTRR279 pKa = 11.84DD280 pKa = 3.48DD281 pKa = 4.17PDD283 pKa = 3.33NGRR286 pKa = 11.84PTCRR290 pKa = 11.84YY291 pKa = 9.17GNCEE295 pKa = 4.14PDD297 pKa = 3.11EE298 pKa = 4.24CLCRR302 pKa = 11.84SNDD305 pKa = 3.88GSDD308 pKa = 3.29EE309 pKa = 4.79LYY311 pKa = 10.95DD312 pKa = 4.07SEE314 pKa = 4.83
MM1 pKa = 6.46TTRR4 pKa = 11.84IRR6 pKa = 11.84AWCFTLNNPTDD17 pKa = 3.66RR18 pKa = 11.84DD19 pKa = 3.54KK20 pKa = 11.43RR21 pKa = 11.84ALIALANTTRR31 pKa = 11.84YY32 pKa = 10.22VIAQLEE38 pKa = 4.3HH39 pKa = 6.95APTTGTPHH47 pKa = 5.39YY48 pKa = 8.8QGYY51 pKa = 7.88MVFHH55 pKa = 7.6DD56 pKa = 4.58GKK58 pKa = 11.0SFSALHH64 pKa = 6.09KK65 pKa = 10.59AMPRR69 pKa = 11.84AHH71 pKa = 6.94FEE73 pKa = 3.97PARR76 pKa = 11.84GTAQQNYY83 pKa = 9.89DD84 pKa = 4.04YY85 pKa = 9.02CTKK88 pKa = 10.63AADD91 pKa = 3.61EE92 pKa = 4.55GQPSNEE98 pKa = 3.81LVLEE102 pKa = 4.64HH103 pKa = 6.49GTLPQQGEE111 pKa = 4.36RR112 pKa = 11.84VDD114 pKa = 3.37ISQVRR119 pKa = 11.84EE120 pKa = 3.89LLRR123 pKa = 11.84SGVTYY128 pKa = 10.79RR129 pKa = 11.84EE130 pKa = 4.09LVDD133 pKa = 3.36MDD135 pKa = 3.9VNYY138 pKa = 10.35QVLRR142 pKa = 11.84IAQEE146 pKa = 3.74WLRR149 pKa = 11.84VHH151 pKa = 6.84EE152 pKa = 4.48PARR155 pKa = 11.84DD156 pKa = 3.85FKK158 pKa = 11.56PEE160 pKa = 3.69VRR162 pKa = 11.84WFYY165 pKa = 11.44GEE167 pKa = 4.04PGAGKK172 pKa = 8.06TRR174 pKa = 11.84AALEE178 pKa = 3.93WLEE181 pKa = 3.9SHH183 pKa = 6.66GEE185 pKa = 4.16VFTVEE190 pKa = 4.33TPAKK194 pKa = 9.98YY195 pKa = 8.51WDD197 pKa = 4.65GYY199 pKa = 10.94DD200 pKa = 3.29GHH202 pKa = 7.14NCVLFDD208 pKa = 5.79DD209 pKa = 5.51LRR211 pKa = 11.84ADD213 pKa = 3.09QCGFVRR219 pKa = 11.84MLRR222 pKa = 11.84LLDD225 pKa = 4.08RR226 pKa = 11.84YY227 pKa = 10.98AMRR230 pKa = 11.84VEE232 pKa = 4.22VKK234 pKa = 10.52GGSKK238 pKa = 8.18QLRR241 pKa = 11.84ARR243 pKa = 11.84YY244 pKa = 8.81IAITAPHH251 pKa = 6.83RR252 pKa = 11.84PEE254 pKa = 4.05EE255 pKa = 4.16IYY257 pKa = 9.43RR258 pKa = 11.84TSSEE262 pKa = 4.42NIVQLTRR269 pKa = 11.84RR270 pKa = 11.84ISSVTHH276 pKa = 4.73VTRR279 pKa = 11.84DD280 pKa = 3.48DD281 pKa = 4.17PDD283 pKa = 3.33NGRR286 pKa = 11.84PTCRR290 pKa = 11.84YY291 pKa = 9.17GNCEE295 pKa = 4.14PDD297 pKa = 3.11EE298 pKa = 4.24CLCRR302 pKa = 11.84SNDD305 pKa = 3.88GSDD308 pKa = 3.29EE309 pKa = 4.79LYY311 pKa = 10.95DD312 pKa = 4.07SEE314 pKa = 4.83
Molecular weight: 36.06 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A190WHC6|A0A190WHC6_9CIRC Uncharacterized protein OS=Circovirus-like genome DCCV-10 OX=1788438 PE=4 SV=1
MM1 pKa = 7.58LQRR4 pKa = 11.84QVHH7 pKa = 4.77RR8 pKa = 11.84TIKK11 pKa = 8.78VTWSSTTVSRR21 pKa = 11.84LVHH24 pKa = 5.23CTRR27 pKa = 11.84RR28 pKa = 11.84CQEE31 pKa = 4.93PISSLPGARR40 pKa = 11.84RR41 pKa = 11.84SKK43 pKa = 10.91IMIIAQRR50 pKa = 11.84LQTKK54 pKa = 9.9ASPPTSSYY62 pKa = 11.17SSTVPYY68 pKa = 9.74PSRR71 pKa = 11.84GRR73 pKa = 11.84GSIYY77 pKa = 9.58PRR79 pKa = 11.84CVSYY83 pKa = 10.64SAPGSHH89 pKa = 6.02IVNWLTWMSITRR101 pKa = 11.84SS102 pKa = 3.42
MM1 pKa = 7.58LQRR4 pKa = 11.84QVHH7 pKa = 4.77RR8 pKa = 11.84TIKK11 pKa = 8.78VTWSSTTVSRR21 pKa = 11.84LVHH24 pKa = 5.23CTRR27 pKa = 11.84RR28 pKa = 11.84CQEE31 pKa = 4.93PISSLPGARR40 pKa = 11.84RR41 pKa = 11.84SKK43 pKa = 10.91IMIIAQRR50 pKa = 11.84LQTKK54 pKa = 9.9ASPPTSSYY62 pKa = 11.17SSTVPYY68 pKa = 9.74PSRR71 pKa = 11.84GRR73 pKa = 11.84GSIYY77 pKa = 9.58PRR79 pKa = 11.84CVSYY83 pKa = 10.64SAPGSHH89 pKa = 6.02IVNWLTWMSITRR101 pKa = 11.84SS102 pKa = 3.42
Molecular weight: 11.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
806 |
102 |
390 |
268.7 |
30.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.444 ± 0.533 | 1.613 ± 0.632 |
5.955 ± 0.845 | 4.218 ± 1.602 |
2.854 ± 0.465 | 5.583 ± 0.363 |
1.985 ± 0.89 | 4.094 ± 0.556 |
5.087 ± 1.382 | 7.32 ± 0.344 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.481 ± 0.27 | 3.846 ± 0.583 |
5.211 ± 0.405 | 4.467 ± 0.149 |
8.437 ± 1.136 | 8.065 ± 1.748 |
9.057 ± 1.024 | 7.072 ± 0.389 |
1.489 ± 0.27 | 3.722 ± 0.573 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
