
Mumia flava
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Nocardioidaceae; Mumia
Average proteome isoelectric point is 5.85
Get precalculated fractions of proteins
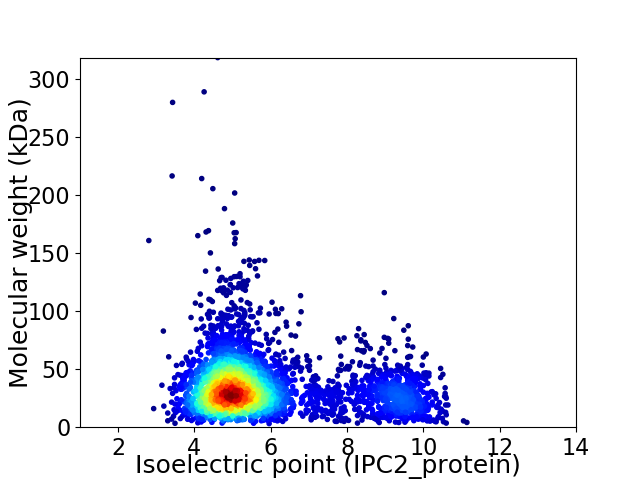
Virtual 2D-PAGE plot for 4037 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2M9BHD8|A0A2M9BHD8_9ACTN Flp pilus assembly protein TadB OS=Mumia flava OX=1348852 GN=CLV56_1596 PE=4 SV=1
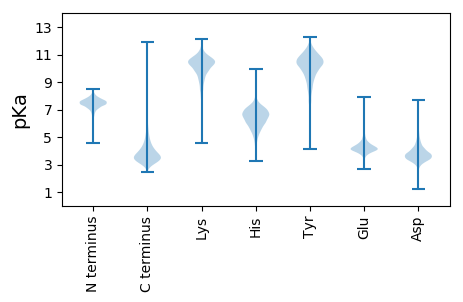
MM1 pKa = 7.36GVADD5 pKa = 5.44DD6 pKa = 4.66EE7 pKa = 4.79GTHH10 pKa = 4.94VRR12 pKa = 11.84GRR14 pKa = 11.84RR15 pKa = 11.84LAVLAAATAILVAGCTSAQDD35 pKa = 3.49ARR37 pKa = 11.84GGDD40 pKa = 4.17DD41 pKa = 3.27PTAAATSSAADD52 pKa = 3.47VRR54 pKa = 11.84TPPDD58 pKa = 3.61PALAPYY64 pKa = 10.33YY65 pKa = 8.8EE66 pKa = 4.23QQVDD70 pKa = 3.03WRR72 pKa = 11.84SCDD75 pKa = 3.23GDD77 pKa = 3.36EE78 pKa = 4.53CADD81 pKa = 3.58VEE83 pKa = 5.28VPLDD87 pKa = 3.53YY88 pKa = 11.27ANPDD92 pKa = 3.43GEE94 pKa = 4.68RR95 pKa = 11.84IALRR99 pKa = 11.84LRR101 pKa = 11.84KK102 pKa = 9.17HH103 pKa = 4.78VAGRR107 pKa = 11.84PDD109 pKa = 3.72DD110 pKa = 3.96RR111 pKa = 11.84VGTLFVNPGGPGVSGVDD128 pKa = 3.32YY129 pKa = 10.58AAYY132 pKa = 9.14FPRR135 pKa = 11.84VAGEE139 pKa = 3.92AVTDD143 pKa = 3.83RR144 pKa = 11.84FDD146 pKa = 4.01VVGFDD151 pKa = 3.66PRR153 pKa = 11.84GVGASSALDD162 pKa = 3.72CGDD165 pKa = 3.47TAEE168 pKa = 3.93VDD170 pKa = 3.81AYY172 pKa = 11.48VEE174 pKa = 4.19TDD176 pKa = 3.35QTPDD180 pKa = 3.49DD181 pKa = 4.38ASEE184 pKa = 3.95EE185 pKa = 4.11QALVEE190 pKa = 4.2ASQTLGEE197 pKa = 4.08EE198 pKa = 4.65CEE200 pKa = 4.29RR201 pKa = 11.84GSGALASHH209 pKa = 6.8VSTVEE214 pKa = 3.47AARR217 pKa = 11.84DD218 pKa = 3.61LDD220 pKa = 3.74VLRR223 pKa = 11.84AVVDD227 pKa = 4.18DD228 pKa = 3.92QALHH232 pKa = 5.89YY233 pKa = 10.61LGASYY238 pKa = 8.2GTKK241 pKa = 10.42LGATYY246 pKa = 11.13AEE248 pKa = 4.48LFADD252 pKa = 3.69RR253 pKa = 11.84VGRR256 pKa = 11.84MVLDD260 pKa = 3.98GAMDD264 pKa = 3.65PTLDD268 pKa = 4.07SEE270 pKa = 4.91GVNLGQAEE278 pKa = 4.58GFQTALEE285 pKa = 4.63AYY287 pKa = 10.03VADD290 pKa = 4.02CVEE293 pKa = 4.31TAGCPLGEE301 pKa = 4.53DD302 pKa = 3.21TDD304 pKa = 4.36AGLEE308 pKa = 4.14EE309 pKa = 5.38IGDD312 pKa = 4.72LIASLDD318 pKa = 4.23DD319 pKa = 5.01DD320 pKa = 5.11PLPTSDD326 pKa = 4.23PDD328 pKa = 3.62RR329 pKa = 11.84PLTEE333 pKa = 4.11ALGFYY338 pKa = 10.44AVAYY342 pKa = 8.48PLYY345 pKa = 10.66NEE347 pKa = 4.81ASWPVLTEE355 pKa = 3.83ALADD359 pKa = 4.1GLDD362 pKa = 3.86GDD364 pKa = 4.78GDD366 pKa = 4.0TMLFVADD373 pKa = 3.96QYY375 pKa = 11.59LSRR378 pKa = 11.84GSDD381 pKa = 3.18GYY383 pKa = 9.79TDD385 pKa = 3.44NSTVAFPAISCLDD398 pKa = 3.48DD399 pKa = 3.81TGDD402 pKa = 3.37VSLDD406 pKa = 3.41EE407 pKa = 4.78ARR409 pKa = 11.84EE410 pKa = 4.0SLAEE414 pKa = 4.05FEE416 pKa = 5.05DD417 pKa = 4.04VSPVLGRR424 pKa = 11.84SFAWSAVGCSTWPMEE439 pKa = 4.34ATEE442 pKa = 4.46PAPEE446 pKa = 4.2IDD448 pKa = 3.91AAGAPPILVVGTTRR462 pKa = 11.84DD463 pKa = 3.33PATPYY468 pKa = 10.89AWAEE472 pKa = 3.9ALADD476 pKa = 3.8QLDD479 pKa = 3.97SGVLVTRR486 pKa = 11.84DD487 pKa = 3.39GDD489 pKa = 3.76GHH491 pKa = 5.96TGYY494 pKa = 10.91AAGNGCVDD502 pKa = 3.65AAVDD506 pKa = 4.02EE507 pKa = 4.43YY508 pKa = 11.47LVDD511 pKa = 3.66GTVPEE516 pKa = 5.66DD517 pKa = 3.72GLEE520 pKa = 4.12CC521 pKa = 5.3
MM1 pKa = 7.36GVADD5 pKa = 5.44DD6 pKa = 4.66EE7 pKa = 4.79GTHH10 pKa = 4.94VRR12 pKa = 11.84GRR14 pKa = 11.84RR15 pKa = 11.84LAVLAAATAILVAGCTSAQDD35 pKa = 3.49ARR37 pKa = 11.84GGDD40 pKa = 4.17DD41 pKa = 3.27PTAAATSSAADD52 pKa = 3.47VRR54 pKa = 11.84TPPDD58 pKa = 3.61PALAPYY64 pKa = 10.33YY65 pKa = 8.8EE66 pKa = 4.23QQVDD70 pKa = 3.03WRR72 pKa = 11.84SCDD75 pKa = 3.23GDD77 pKa = 3.36EE78 pKa = 4.53CADD81 pKa = 3.58VEE83 pKa = 5.28VPLDD87 pKa = 3.53YY88 pKa = 11.27ANPDD92 pKa = 3.43GEE94 pKa = 4.68RR95 pKa = 11.84IALRR99 pKa = 11.84LRR101 pKa = 11.84KK102 pKa = 9.17HH103 pKa = 4.78VAGRR107 pKa = 11.84PDD109 pKa = 3.72DD110 pKa = 3.96RR111 pKa = 11.84VGTLFVNPGGPGVSGVDD128 pKa = 3.32YY129 pKa = 10.58AAYY132 pKa = 9.14FPRR135 pKa = 11.84VAGEE139 pKa = 3.92AVTDD143 pKa = 3.83RR144 pKa = 11.84FDD146 pKa = 4.01VVGFDD151 pKa = 3.66PRR153 pKa = 11.84GVGASSALDD162 pKa = 3.72CGDD165 pKa = 3.47TAEE168 pKa = 3.93VDD170 pKa = 3.81AYY172 pKa = 11.48VEE174 pKa = 4.19TDD176 pKa = 3.35QTPDD180 pKa = 3.49DD181 pKa = 4.38ASEE184 pKa = 3.95EE185 pKa = 4.11QALVEE190 pKa = 4.2ASQTLGEE197 pKa = 4.08EE198 pKa = 4.65CEE200 pKa = 4.29RR201 pKa = 11.84GSGALASHH209 pKa = 6.8VSTVEE214 pKa = 3.47AARR217 pKa = 11.84DD218 pKa = 3.61LDD220 pKa = 3.74VLRR223 pKa = 11.84AVVDD227 pKa = 4.18DD228 pKa = 3.92QALHH232 pKa = 5.89YY233 pKa = 10.61LGASYY238 pKa = 8.2GTKK241 pKa = 10.42LGATYY246 pKa = 11.13AEE248 pKa = 4.48LFADD252 pKa = 3.69RR253 pKa = 11.84VGRR256 pKa = 11.84MVLDD260 pKa = 3.98GAMDD264 pKa = 3.65PTLDD268 pKa = 4.07SEE270 pKa = 4.91GVNLGQAEE278 pKa = 4.58GFQTALEE285 pKa = 4.63AYY287 pKa = 10.03VADD290 pKa = 4.02CVEE293 pKa = 4.31TAGCPLGEE301 pKa = 4.53DD302 pKa = 3.21TDD304 pKa = 4.36AGLEE308 pKa = 4.14EE309 pKa = 5.38IGDD312 pKa = 4.72LIASLDD318 pKa = 4.23DD319 pKa = 5.01DD320 pKa = 5.11PLPTSDD326 pKa = 4.23PDD328 pKa = 3.62RR329 pKa = 11.84PLTEE333 pKa = 4.11ALGFYY338 pKa = 10.44AVAYY342 pKa = 8.48PLYY345 pKa = 10.66NEE347 pKa = 4.81ASWPVLTEE355 pKa = 3.83ALADD359 pKa = 4.1GLDD362 pKa = 3.86GDD364 pKa = 4.78GDD366 pKa = 4.0TMLFVADD373 pKa = 3.96QYY375 pKa = 11.59LSRR378 pKa = 11.84GSDD381 pKa = 3.18GYY383 pKa = 9.79TDD385 pKa = 3.44NSTVAFPAISCLDD398 pKa = 3.48DD399 pKa = 3.81TGDD402 pKa = 3.37VSLDD406 pKa = 3.41EE407 pKa = 4.78ARR409 pKa = 11.84EE410 pKa = 4.0SLAEE414 pKa = 4.05FEE416 pKa = 5.05DD417 pKa = 4.04VSPVLGRR424 pKa = 11.84SFAWSAVGCSTWPMEE439 pKa = 4.34ATEE442 pKa = 4.46PAPEE446 pKa = 4.2IDD448 pKa = 3.91AAGAPPILVVGTTRR462 pKa = 11.84DD463 pKa = 3.33PATPYY468 pKa = 10.89AWAEE472 pKa = 3.9ALADD476 pKa = 3.8QLDD479 pKa = 3.97SGVLVTRR486 pKa = 11.84DD487 pKa = 3.39GDD489 pKa = 3.76GHH491 pKa = 5.96TGYY494 pKa = 10.91AAGNGCVDD502 pKa = 3.65AAVDD506 pKa = 4.02EE507 pKa = 4.43YY508 pKa = 11.47LVDD511 pKa = 3.66GTVPEE516 pKa = 5.66DD517 pKa = 3.72GLEE520 pKa = 4.12CC521 pKa = 5.3
Molecular weight: 54.24 kDa
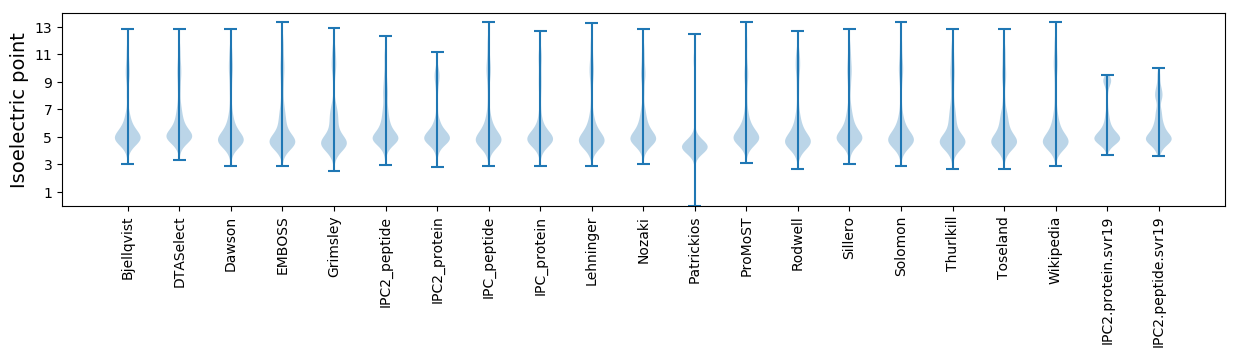
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2M9BKK2|A0A2M9BKK2_9ACTN Succinate dehydrogenase / fumarate reductase flavoprotein subunit OS=Mumia flava OX=1348852 GN=CLV56_2715 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.38KK7 pKa = 8.42RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.06RR11 pKa = 11.84MSKK14 pKa = 9.76KK15 pKa = 9.68KK16 pKa = 9.77HH17 pKa = 5.59RR18 pKa = 11.84KK19 pKa = 7.37LLKK22 pKa = 8.07RR23 pKa = 11.84TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 10.11LGKK33 pKa = 9.87
MM1 pKa = 7.4GSVIKK6 pKa = 10.38KK7 pKa = 8.42RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.06RR11 pKa = 11.84MSKK14 pKa = 9.76KK15 pKa = 9.68KK16 pKa = 9.77HH17 pKa = 5.59RR18 pKa = 11.84KK19 pKa = 7.37LLKK22 pKa = 8.07RR23 pKa = 11.84TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 10.11LGKK33 pKa = 9.87
Molecular weight: 4.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1354464 |
29 |
3061 |
335.5 |
35.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.653 ± 0.059 | 0.665 ± 0.01 |
6.954 ± 0.038 | 5.65 ± 0.033 |
2.662 ± 0.023 | 9.224 ± 0.032 |
2.036 ± 0.018 | 3.392 ± 0.025 |
1.612 ± 0.03 | 9.998 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.756 ± 0.013 | 1.567 ± 0.021 |
5.634 ± 0.031 | 2.492 ± 0.021 |
7.971 ± 0.047 | 5.294 ± 0.026 |
6.214 ± 0.042 | 9.685 ± 0.039 |
1.514 ± 0.017 | 2.025 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
