
Polaribacter sp. Hel1_33_78
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Polaribacter; unclassified Polaribacter
Average proteome isoelectric point is 7.04
Get precalculated fractions of proteins
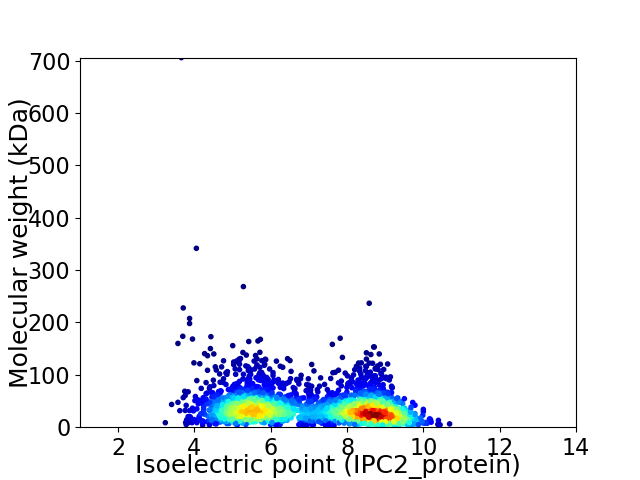
Virtual 2D-PAGE plot for 2741 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H2DZJ2|A0A1H2DZJ2_9FLAO Nucleotide-binding universal stress protein UspA family OS=Polaribacter sp. Hel1_33_78 OX=1336804 GN=SAMN04487762_0233 PE=3 SV=1
MM1 pKa = 7.7LKK3 pKa = 9.19TFKK6 pKa = 10.47TIIFLFLFSLVFSCSPDD23 pKa = 2.96EE24 pKa = 4.43KK25 pKa = 11.23VNNDD29 pKa = 3.57RR30 pKa = 11.84DD31 pKa = 3.62DD32 pKa = 4.07DD33 pKa = 4.45QIVNSSDD40 pKa = 3.37NCPDD44 pKa = 3.57TQNEE48 pKa = 4.16DD49 pKa = 3.76QLDD52 pKa = 3.82SDD54 pKa = 4.16NDD56 pKa = 4.92GIGDD60 pKa = 4.4LCDD63 pKa = 5.87DD64 pKa = 5.33DD65 pKa = 6.61DD66 pKa = 6.99DD67 pKa = 5.99NDD69 pKa = 4.3GVLDD73 pKa = 4.05TDD75 pKa = 4.88DD76 pKa = 4.53NCPLIANPAQTDD88 pKa = 3.04VDD90 pKa = 3.8NDD92 pKa = 4.09GKK94 pKa = 11.49GDD96 pKa = 3.62TCDD99 pKa = 4.82NDD101 pKa = 5.91DD102 pKa = 5.54DD103 pKa = 6.61DD104 pKa = 6.49SDD106 pKa = 4.21NDD108 pKa = 4.73GIKK111 pKa = 10.56DD112 pKa = 4.65SEE114 pKa = 4.75DD115 pKa = 3.32NCPNIANPNQEE126 pKa = 4.72DD127 pKa = 4.17EE128 pKa = 4.5NNDD131 pKa = 4.24GIGDD135 pKa = 4.16ACASTNKK142 pKa = 9.27FVCEE146 pKa = 3.66NGIANGYY153 pKa = 7.9PCNDD157 pKa = 3.26YY158 pKa = 11.44DD159 pKa = 5.1LLLNIPLATFSASAGNDD176 pKa = 2.8SWGWTDD182 pKa = 3.13ATNGKK187 pKa = 9.43EE188 pKa = 3.87YY189 pKa = 10.78AIMGLNNGTAFIDD202 pKa = 3.51ITDD205 pKa = 4.03TEE207 pKa = 4.4NPVYY211 pKa = 10.42LGKK214 pKa = 10.83LPTASVNSSWRR225 pKa = 11.84DD226 pKa = 2.96IKK228 pKa = 11.06VYY230 pKa = 10.21KK231 pKa = 10.5DD232 pKa = 2.88HH233 pKa = 7.13AFIVSEE239 pKa = 4.02ASNHH243 pKa = 4.45GMQVFDD249 pKa = 3.68LTRR252 pKa = 11.84LRR254 pKa = 11.84NVSNTPATFTADD266 pKa = 3.2TNFTEE271 pKa = 4.91FGKK274 pKa = 10.64AHH276 pKa = 6.61NIVINEE282 pKa = 4.1TSGYY286 pKa = 9.95AYY288 pKa = 9.69IVGASGNSTYY298 pKa = 11.12GGGPIFINIQDD309 pKa = 3.79PKK311 pKa = 10.92NPVSEE316 pKa = 4.56GGFSEE321 pKa = 4.91GGYY324 pKa = 9.66SHH326 pKa = 7.9DD327 pKa = 4.0AQVITYY333 pKa = 8.64NGPDD337 pKa = 3.16TDD339 pKa = 4.06YY340 pKa = 10.45TGKK343 pKa = 10.72EE344 pKa = 3.49ILIGSNEE351 pKa = 4.12NEE353 pKa = 4.15VVIADD358 pKa = 3.82ITDD361 pKa = 3.35KK362 pKa = 8.51TTPTIISTISYY373 pKa = 10.52SNVAYY378 pKa = 10.23AHH380 pKa = 5.84QGWFTEE386 pKa = 3.98DD387 pKa = 2.81LKK389 pKa = 11.63YY390 pKa = 10.73FILGDD395 pKa = 3.79EE396 pKa = 4.18LDD398 pKa = 4.19EE399 pKa = 6.09RR400 pKa = 11.84DD401 pKa = 3.7FGNNTRR407 pKa = 11.84SIIFDD412 pKa = 3.52FTDD415 pKa = 3.74LDD417 pKa = 4.07NPSHH421 pKa = 6.49HH422 pKa = 7.08FDD424 pKa = 3.66YY425 pKa = 10.99FGNTAAIDD433 pKa = 3.38HH434 pKa = 6.38NGYY437 pKa = 10.07VKK439 pKa = 10.8EE440 pKa = 4.13NIYY443 pKa = 10.53YY444 pKa = 8.55QANYY448 pKa = 8.03TAGVRR453 pKa = 11.84MIDD456 pKa = 4.34VSNGDD461 pKa = 3.11NKK463 pKa = 11.08DD464 pKa = 3.62FNEE467 pKa = 3.96VGFFDD472 pKa = 4.92TYY474 pKa = 11.3PNNNNTAFNGVWNVYY489 pKa = 10.19PYY491 pKa = 10.29FPSGNIIISDD501 pKa = 3.38IDD503 pKa = 3.34NGFFVIKK510 pKa = 10.25KK511 pKa = 9.81SNPP514 pKa = 3.15
MM1 pKa = 7.7LKK3 pKa = 9.19TFKK6 pKa = 10.47TIIFLFLFSLVFSCSPDD23 pKa = 2.96EE24 pKa = 4.43KK25 pKa = 11.23VNNDD29 pKa = 3.57RR30 pKa = 11.84DD31 pKa = 3.62DD32 pKa = 4.07DD33 pKa = 4.45QIVNSSDD40 pKa = 3.37NCPDD44 pKa = 3.57TQNEE48 pKa = 4.16DD49 pKa = 3.76QLDD52 pKa = 3.82SDD54 pKa = 4.16NDD56 pKa = 4.92GIGDD60 pKa = 4.4LCDD63 pKa = 5.87DD64 pKa = 5.33DD65 pKa = 6.61DD66 pKa = 6.99DD67 pKa = 5.99NDD69 pKa = 4.3GVLDD73 pKa = 4.05TDD75 pKa = 4.88DD76 pKa = 4.53NCPLIANPAQTDD88 pKa = 3.04VDD90 pKa = 3.8NDD92 pKa = 4.09GKK94 pKa = 11.49GDD96 pKa = 3.62TCDD99 pKa = 4.82NDD101 pKa = 5.91DD102 pKa = 5.54DD103 pKa = 6.61DD104 pKa = 6.49SDD106 pKa = 4.21NDD108 pKa = 4.73GIKK111 pKa = 10.56DD112 pKa = 4.65SEE114 pKa = 4.75DD115 pKa = 3.32NCPNIANPNQEE126 pKa = 4.72DD127 pKa = 4.17EE128 pKa = 4.5NNDD131 pKa = 4.24GIGDD135 pKa = 4.16ACASTNKK142 pKa = 9.27FVCEE146 pKa = 3.66NGIANGYY153 pKa = 7.9PCNDD157 pKa = 3.26YY158 pKa = 11.44DD159 pKa = 5.1LLLNIPLATFSASAGNDD176 pKa = 2.8SWGWTDD182 pKa = 3.13ATNGKK187 pKa = 9.43EE188 pKa = 3.87YY189 pKa = 10.78AIMGLNNGTAFIDD202 pKa = 3.51ITDD205 pKa = 4.03TEE207 pKa = 4.4NPVYY211 pKa = 10.42LGKK214 pKa = 10.83LPTASVNSSWRR225 pKa = 11.84DD226 pKa = 2.96IKK228 pKa = 11.06VYY230 pKa = 10.21KK231 pKa = 10.5DD232 pKa = 2.88HH233 pKa = 7.13AFIVSEE239 pKa = 4.02ASNHH243 pKa = 4.45GMQVFDD249 pKa = 3.68LTRR252 pKa = 11.84LRR254 pKa = 11.84NVSNTPATFTADD266 pKa = 3.2TNFTEE271 pKa = 4.91FGKK274 pKa = 10.64AHH276 pKa = 6.61NIVINEE282 pKa = 4.1TSGYY286 pKa = 9.95AYY288 pKa = 9.69IVGASGNSTYY298 pKa = 11.12GGGPIFINIQDD309 pKa = 3.79PKK311 pKa = 10.92NPVSEE316 pKa = 4.56GGFSEE321 pKa = 4.91GGYY324 pKa = 9.66SHH326 pKa = 7.9DD327 pKa = 4.0AQVITYY333 pKa = 8.64NGPDD337 pKa = 3.16TDD339 pKa = 4.06YY340 pKa = 10.45TGKK343 pKa = 10.72EE344 pKa = 3.49ILIGSNEE351 pKa = 4.12NEE353 pKa = 4.15VVIADD358 pKa = 3.82ITDD361 pKa = 3.35KK362 pKa = 8.51TTPTIISTISYY373 pKa = 10.52SNVAYY378 pKa = 10.23AHH380 pKa = 5.84QGWFTEE386 pKa = 3.98DD387 pKa = 2.81LKK389 pKa = 11.63YY390 pKa = 10.73FILGDD395 pKa = 3.79EE396 pKa = 4.18LDD398 pKa = 4.19EE399 pKa = 6.09RR400 pKa = 11.84DD401 pKa = 3.7FGNNTRR407 pKa = 11.84SIIFDD412 pKa = 3.52FTDD415 pKa = 3.74LDD417 pKa = 4.07NPSHH421 pKa = 6.49HH422 pKa = 7.08FDD424 pKa = 3.66YY425 pKa = 10.99FGNTAAIDD433 pKa = 3.38HH434 pKa = 6.38NGYY437 pKa = 10.07VKK439 pKa = 10.8EE440 pKa = 4.13NIYY443 pKa = 10.53YY444 pKa = 8.55QANYY448 pKa = 8.03TAGVRR453 pKa = 11.84MIDD456 pKa = 4.34VSNGDD461 pKa = 3.11NKK463 pKa = 11.08DD464 pKa = 3.62FNEE467 pKa = 3.96VGFFDD472 pKa = 4.92TYY474 pKa = 11.3PNNNNTAFNGVWNVYY489 pKa = 10.19PYY491 pKa = 10.29FPSGNIIISDD501 pKa = 3.38IDD503 pKa = 3.34NGFFVIKK510 pKa = 10.25KK511 pKa = 9.81SNPP514 pKa = 3.15
Molecular weight: 56.46 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H2ESI0|A0A1H2ESI0_9FLAO CarboxypepD_reg-like domain-containing protein OS=Polaribacter sp. Hel1_33_78 OX=1336804 GN=SAMN04487762_1060 PE=4 SV=1
MM1 pKa = 7.2NFIHH5 pKa = 7.2AVRR8 pKa = 11.84HH9 pKa = 4.49FFEE12 pKa = 4.16RR13 pKa = 11.84HH14 pKa = 4.23GFGVFSRR21 pKa = 11.84FADD24 pKa = 3.64KK25 pKa = 11.22LGMRR29 pKa = 11.84AKK31 pKa = 10.16NVRR34 pKa = 11.84IFFIYY39 pKa = 10.2ISFVTVGLSFGLYY52 pKa = 8.32LTLAFLFRR60 pKa = 11.84LKK62 pKa = 11.25DD63 pKa = 3.32MIYY66 pKa = 9.31TKK68 pKa = 10.37RR69 pKa = 11.84SSVFDD74 pKa = 3.68LL75 pKa = 4.35
MM1 pKa = 7.2NFIHH5 pKa = 7.2AVRR8 pKa = 11.84HH9 pKa = 4.49FFEE12 pKa = 4.16RR13 pKa = 11.84HH14 pKa = 4.23GFGVFSRR21 pKa = 11.84FADD24 pKa = 3.64KK25 pKa = 11.22LGMRR29 pKa = 11.84AKK31 pKa = 10.16NVRR34 pKa = 11.84IFFIYY39 pKa = 10.2ISFVTVGLSFGLYY52 pKa = 8.32LTLAFLFRR60 pKa = 11.84LKK62 pKa = 11.25DD63 pKa = 3.32MIYY66 pKa = 9.31TKK68 pKa = 10.37RR69 pKa = 11.84SSVFDD74 pKa = 3.68LL75 pKa = 4.35
Molecular weight: 8.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
952477 |
29 |
6769 |
347.5 |
39.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.071 ± 0.045 | 0.705 ± 0.014 |
5.34 ± 0.046 | 6.446 ± 0.051 |
5.527 ± 0.048 | 6.319 ± 0.054 |
1.693 ± 0.021 | 8.641 ± 0.05 |
8.696 ± 0.069 | 9.086 ± 0.063 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.09 ± 0.028 | 6.619 ± 0.053 |
3.111 ± 0.022 | 3.175 ± 0.028 |
3.177 ± 0.028 | 6.66 ± 0.045 |
5.809 ± 0.079 | 5.951 ± 0.036 |
1.028 ± 0.017 | 3.855 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
