
Corchorus yellow vein virus - [Hoa Binh]
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Begomovirus; Corchorus yellow vein virus
Average proteome isoelectric point is 8.3
Get precalculated fractions of proteins
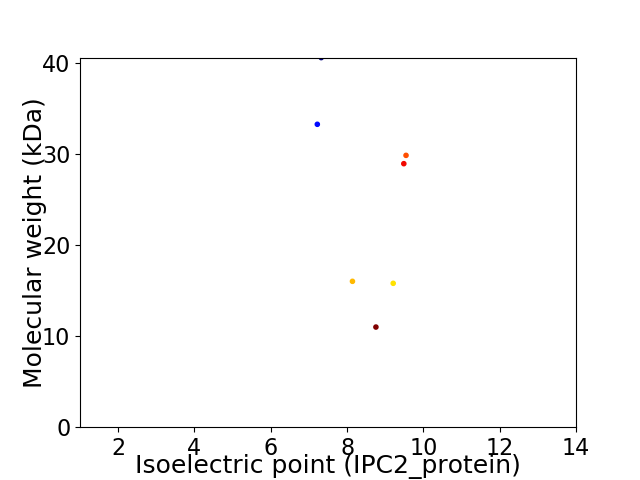
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q645G8|Q645G8_9GEMI Nuclear shuttle protein OS=Corchorus yellow vein virus - [Hoa Binh] OX=293284 GN=BV1 PE=3 SV=1
MM1 pKa = 7.74EE2 pKa = 4.83SQLAQAPSSFNYY14 pKa = 9.76IEE16 pKa = 4.09SHH18 pKa = 6.17RR19 pKa = 11.84DD20 pKa = 3.25EE21 pKa = 4.44YY22 pKa = 11.48RR23 pKa = 11.84LTHH26 pKa = 7.07DD27 pKa = 3.36LTEE30 pKa = 4.44IVLQFPSTAEE40 pKa = 3.5QWGATVRR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84CMKK52 pKa = 9.72IDD54 pKa = 3.27HH55 pKa = 6.48CVIEE59 pKa = 4.31YY60 pKa = 9.79RR61 pKa = 11.84QQVPINATGSVIVEE75 pKa = 3.82IHH77 pKa = 7.1DD78 pKa = 4.31KK79 pKa = 11.26RR80 pKa = 11.84MTDD83 pKa = 3.41NEE85 pKa = 4.46SLQASYY91 pKa = 9.77TFPIRR96 pKa = 11.84CNIDD100 pKa = 2.86LHH102 pKa = 6.12YY103 pKa = 10.57FSASFFSLKK112 pKa = 10.83DD113 pKa = 3.77PIPWKK118 pKa = 10.22LYY120 pKa = 10.12YY121 pKa = 9.87RR122 pKa = 11.84VSDD125 pKa = 3.88TNVHH129 pKa = 5.9QGTHH133 pKa = 4.64FAKK136 pKa = 10.88FKK138 pKa = 11.0GKK140 pKa = 10.75LKK142 pKa = 10.36MSTAKK147 pKa = 10.51HH148 pKa = 5.34SVDD151 pKa = 3.66VIFRR155 pKa = 11.84SPTVKK160 pKa = 10.15ILSKK164 pKa = 10.78QFTGKK169 pKa = 10.37DD170 pKa = 2.79IDD172 pKa = 4.26FSHH175 pKa = 7.02VDD177 pKa = 3.27YY178 pKa = 11.46GKK180 pKa = 10.29VEE182 pKa = 4.18RR183 pKa = 11.84KK184 pKa = 9.14LVKK187 pKa = 10.4CEE189 pKa = 3.68SSSRR193 pKa = 11.84LGLHH197 pKa = 5.88SAIEE201 pKa = 4.02LRR203 pKa = 11.84PGEE206 pKa = 4.2SWATRR211 pKa = 11.84SCIGPDD217 pKa = 3.12TMEE220 pKa = 4.52TEE222 pKa = 4.46SEE224 pKa = 4.3VHH226 pKa = 5.96NRR228 pKa = 11.84NHH230 pKa = 7.86PYY232 pKa = 10.35RR233 pKa = 11.84EE234 pKa = 4.15LNRR237 pKa = 11.84LSTSMLDD244 pKa = 3.84PGDD247 pKa = 3.96SASMIGASRR256 pKa = 11.84AEE258 pKa = 4.24SNITLTRR265 pKa = 11.84AQLQEE270 pKa = 4.35LVSSTVEE277 pKa = 3.45HH278 pKa = 6.95CINRR282 pKa = 11.84NCTPHH287 pKa = 5.59QAKK290 pKa = 9.86PLL292 pKa = 3.84
MM1 pKa = 7.74EE2 pKa = 4.83SQLAQAPSSFNYY14 pKa = 9.76IEE16 pKa = 4.09SHH18 pKa = 6.17RR19 pKa = 11.84DD20 pKa = 3.25EE21 pKa = 4.44YY22 pKa = 11.48RR23 pKa = 11.84LTHH26 pKa = 7.07DD27 pKa = 3.36LTEE30 pKa = 4.44IVLQFPSTAEE40 pKa = 3.5QWGATVRR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84CMKK52 pKa = 9.72IDD54 pKa = 3.27HH55 pKa = 6.48CVIEE59 pKa = 4.31YY60 pKa = 9.79RR61 pKa = 11.84QQVPINATGSVIVEE75 pKa = 3.82IHH77 pKa = 7.1DD78 pKa = 4.31KK79 pKa = 11.26RR80 pKa = 11.84MTDD83 pKa = 3.41NEE85 pKa = 4.46SLQASYY91 pKa = 9.77TFPIRR96 pKa = 11.84CNIDD100 pKa = 2.86LHH102 pKa = 6.12YY103 pKa = 10.57FSASFFSLKK112 pKa = 10.83DD113 pKa = 3.77PIPWKK118 pKa = 10.22LYY120 pKa = 10.12YY121 pKa = 9.87RR122 pKa = 11.84VSDD125 pKa = 3.88TNVHH129 pKa = 5.9QGTHH133 pKa = 4.64FAKK136 pKa = 10.88FKK138 pKa = 11.0GKK140 pKa = 10.75LKK142 pKa = 10.36MSTAKK147 pKa = 10.51HH148 pKa = 5.34SVDD151 pKa = 3.66VIFRR155 pKa = 11.84SPTVKK160 pKa = 10.15ILSKK164 pKa = 10.78QFTGKK169 pKa = 10.37DD170 pKa = 2.79IDD172 pKa = 4.26FSHH175 pKa = 7.02VDD177 pKa = 3.27YY178 pKa = 11.46GKK180 pKa = 10.29VEE182 pKa = 4.18RR183 pKa = 11.84KK184 pKa = 9.14LVKK187 pKa = 10.4CEE189 pKa = 3.68SSSRR193 pKa = 11.84LGLHH197 pKa = 5.88SAIEE201 pKa = 4.02LRR203 pKa = 11.84PGEE206 pKa = 4.2SWATRR211 pKa = 11.84SCIGPDD217 pKa = 3.12TMEE220 pKa = 4.52TEE222 pKa = 4.46SEE224 pKa = 4.3VHH226 pKa = 5.96NRR228 pKa = 11.84NHH230 pKa = 7.86PYY232 pKa = 10.35RR233 pKa = 11.84EE234 pKa = 4.15LNRR237 pKa = 11.84LSTSMLDD244 pKa = 3.84PGDD247 pKa = 3.96SASMIGASRR256 pKa = 11.84AEE258 pKa = 4.24SNITLTRR265 pKa = 11.84AQLQEE270 pKa = 4.35LVSSTVEE277 pKa = 3.45HH278 pKa = 6.95CINRR282 pKa = 11.84NCTPHH287 pKa = 5.59QAKK290 pKa = 9.86PLL292 pKa = 3.84
Molecular weight: 33.31 kDa
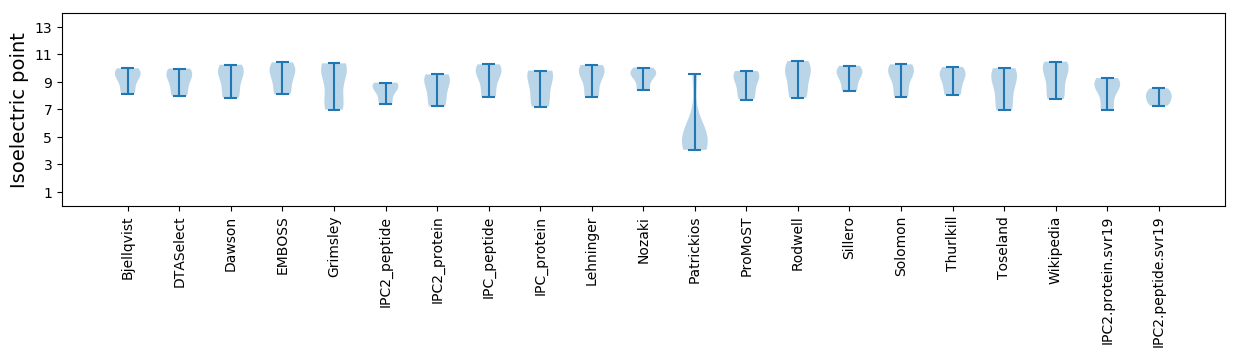
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q645H3|Q645H3_9GEMI Capsid protein OS=Corchorus yellow vein virus - [Hoa Binh] OX=293284 GN=AV1 PE=3 SV=1
MM1 pKa = 7.98PKK3 pKa = 9.65RR4 pKa = 11.84DD5 pKa = 3.82APWRR9 pKa = 11.84LMAGTSKK16 pKa = 10.64VSRR19 pKa = 11.84SSNYY23 pKa = 9.02SPRR26 pKa = 11.84GGVSDD31 pKa = 3.63SGSYY35 pKa = 10.25LPRR38 pKa = 11.84RR39 pKa = 11.84FSRR42 pKa = 11.84ASLWANRR49 pKa = 11.84PMYY52 pKa = 9.88RR53 pKa = 11.84KK54 pKa = 8.83PRR56 pKa = 11.84IYY58 pKa = 9.41RR59 pKa = 11.84TYY61 pKa = 10.72RR62 pKa = 11.84SPDD65 pKa = 3.24VPKK68 pKa = 10.81GCEE71 pKa = 4.39GPCKK75 pKa = 10.23VQSFEE80 pKa = 3.84QRR82 pKa = 11.84HH83 pKa = 6.27DD84 pKa = 3.18IAHH87 pKa = 6.05TGKK90 pKa = 10.57VICLSDD96 pKa = 3.31VTRR99 pKa = 11.84GNGITHH105 pKa = 6.78RR106 pKa = 11.84VGKK109 pKa = 9.44RR110 pKa = 11.84FCVKK114 pKa = 9.92SVYY117 pKa = 10.16ILGKK121 pKa = 9.37IWMDD125 pKa = 3.32EE126 pKa = 4.0NIKK129 pKa = 10.42VKK131 pKa = 10.63NHH133 pKa = 5.61TNSVMFWLVRR143 pKa = 11.84DD144 pKa = 3.83RR145 pKa = 11.84RR146 pKa = 11.84PFGSPMDD153 pKa = 4.16FGQVFNMYY161 pKa = 10.66DD162 pKa = 3.86NEE164 pKa = 4.34PSTATVKK171 pKa = 10.81NDD173 pKa = 2.84LRR175 pKa = 11.84DD176 pKa = 3.6RR177 pKa = 11.84YY178 pKa = 10.31QVMHH182 pKa = 7.01RR183 pKa = 11.84FSAKK187 pKa = 8.35VTGGQYY193 pKa = 11.1ASNEE197 pKa = 3.84QALVRR202 pKa = 11.84RR203 pKa = 11.84FWKK206 pKa = 10.4VNNHH210 pKa = 4.48VVYY213 pKa = 10.66NHH215 pKa = 5.84QEE217 pKa = 3.45AAKK220 pKa = 10.4YY221 pKa = 9.25EE222 pKa = 4.13NHH224 pKa = 6.46TEE226 pKa = 3.99NALLLYY232 pKa = 7.29MACTHH237 pKa = 7.07ASNPVYY243 pKa = 9.86ATLKK247 pKa = 9.47IRR249 pKa = 11.84IYY251 pKa = 10.59FYY253 pKa = 11.32DD254 pKa = 4.06SISNN258 pKa = 3.66
MM1 pKa = 7.98PKK3 pKa = 9.65RR4 pKa = 11.84DD5 pKa = 3.82APWRR9 pKa = 11.84LMAGTSKK16 pKa = 10.64VSRR19 pKa = 11.84SSNYY23 pKa = 9.02SPRR26 pKa = 11.84GGVSDD31 pKa = 3.63SGSYY35 pKa = 10.25LPRR38 pKa = 11.84RR39 pKa = 11.84FSRR42 pKa = 11.84ASLWANRR49 pKa = 11.84PMYY52 pKa = 9.88RR53 pKa = 11.84KK54 pKa = 8.83PRR56 pKa = 11.84IYY58 pKa = 9.41RR59 pKa = 11.84TYY61 pKa = 10.72RR62 pKa = 11.84SPDD65 pKa = 3.24VPKK68 pKa = 10.81GCEE71 pKa = 4.39GPCKK75 pKa = 10.23VQSFEE80 pKa = 3.84QRR82 pKa = 11.84HH83 pKa = 6.27DD84 pKa = 3.18IAHH87 pKa = 6.05TGKK90 pKa = 10.57VICLSDD96 pKa = 3.31VTRR99 pKa = 11.84GNGITHH105 pKa = 6.78RR106 pKa = 11.84VGKK109 pKa = 9.44RR110 pKa = 11.84FCVKK114 pKa = 9.92SVYY117 pKa = 10.16ILGKK121 pKa = 9.37IWMDD125 pKa = 3.32EE126 pKa = 4.0NIKK129 pKa = 10.42VKK131 pKa = 10.63NHH133 pKa = 5.61TNSVMFWLVRR143 pKa = 11.84DD144 pKa = 3.83RR145 pKa = 11.84RR146 pKa = 11.84PFGSPMDD153 pKa = 4.16FGQVFNMYY161 pKa = 10.66DD162 pKa = 3.86NEE164 pKa = 4.34PSTATVKK171 pKa = 10.81NDD173 pKa = 2.84LRR175 pKa = 11.84DD176 pKa = 3.6RR177 pKa = 11.84YY178 pKa = 10.31QVMHH182 pKa = 7.01RR183 pKa = 11.84FSAKK187 pKa = 8.35VTGGQYY193 pKa = 11.1ASNEE197 pKa = 3.84QALVRR202 pKa = 11.84RR203 pKa = 11.84FWKK206 pKa = 10.4VNNHH210 pKa = 4.48VVYY213 pKa = 10.66NHH215 pKa = 5.84QEE217 pKa = 3.45AAKK220 pKa = 10.4YY221 pKa = 9.25EE222 pKa = 4.13NHH224 pKa = 6.46TEE226 pKa = 3.99NALLLYY232 pKa = 7.29MACTHH237 pKa = 7.07ASNPVYY243 pKa = 9.86ATLKK247 pKa = 9.47IRR249 pKa = 11.84IYY251 pKa = 10.59FYY253 pKa = 11.32DD254 pKa = 4.06SISNN258 pKa = 3.66
Molecular weight: 29.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1529 |
96 |
359 |
218.4 |
25.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.494 ± 0.305 | 2.224 ± 0.151 |
4.709 ± 0.238 | 4.709 ± 0.591 |
4.513 ± 0.418 | 4.578 ± 0.484 |
3.597 ± 0.477 | 5.821 ± 0.681 |
5.298 ± 0.563 | 6.475 ± 0.373 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.42 ± 0.466 | 5.755 ± 0.746 |
5.69 ± 0.421 | 3.728 ± 0.43 |
7.848 ± 1.046 | 9.614 ± 0.546 |
6.148 ± 0.423 | 5.559 ± 0.556 |
1.439 ± 0.273 | 4.382 ± 0.546 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
