
Candidatus Marinamargulisbacteria bacterium SCGC AG-414-C22
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Candidatus Margulisbacteria; Candidatus Marinamargulisbacteria
Average proteome isoelectric point is 6.82
Get precalculated fractions of proteins
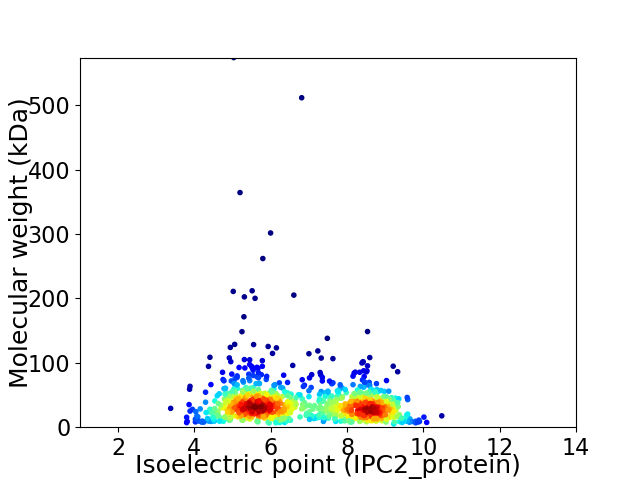
Virtual 2D-PAGE plot for 930 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A328RF38|A0A328RF38_9BACT Uncharacterized protein OS=Candidatus Marinamargulisbacteria bacterium SCGC AG-414-C22 OX=2184344 GN=DID76_00170 PE=4 SV=1
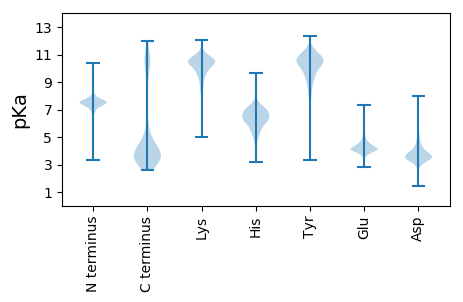
MM1 pKa = 7.45KK2 pKa = 10.32FKK4 pKa = 11.01LFFIIFTITLLVGCSSSEE22 pKa = 3.74VSTEE26 pKa = 3.38NDD28 pKa = 3.1TVYY31 pKa = 11.22EE32 pKa = 3.91NLTFSGHH39 pKa = 4.3VAIDD43 pKa = 3.67ASSGTSSLEE52 pKa = 3.54ASAYY56 pKa = 10.65ALLSNSNAPSTDD68 pKa = 2.74EE69 pKa = 4.11SNYY72 pKa = 10.14MIVIQNQISNRR83 pKa = 11.84LYY85 pKa = 10.89LSQLDD90 pKa = 3.74ADD92 pKa = 4.42GTFHH96 pKa = 7.62FSDD99 pKa = 3.98NGTDD103 pKa = 3.32AQAIQSVNHH112 pKa = 6.62DD113 pKa = 3.45GGYY116 pKa = 10.54GNHH119 pKa = 5.9FMVMLLQKK127 pKa = 10.92DD128 pKa = 3.75PLEE131 pKa = 4.63VKK133 pKa = 10.43AVAYY137 pKa = 10.55LPGTSEE143 pKa = 3.72EE144 pKa = 5.1GYY146 pKa = 10.76SGFQFSEE153 pKa = 4.37SFDD156 pKa = 3.56QEE158 pKa = 3.99LQFNYY163 pKa = 10.59NSDD166 pKa = 3.96SASMTVSEE174 pKa = 4.21THH176 pKa = 6.61INDD179 pKa = 3.28IAGLQVDD186 pKa = 3.69EE187 pKa = 5.15DD188 pKa = 4.27FVVRR192 pKa = 11.84LVNDD196 pKa = 4.34TPAGATNFGKK206 pKa = 8.6STDD209 pKa = 3.79SQTSTVSDD217 pKa = 3.76INQLDD222 pKa = 4.16PDD224 pKa = 3.81QDD226 pKa = 3.73GQPNIFDD233 pKa = 3.82GMNNGSDD240 pKa = 4.4LDD242 pKa = 4.15NLSADD247 pKa = 3.83NKK249 pKa = 10.2TEE251 pKa = 3.86GATVSDD257 pKa = 5.01SIASSIMFMNLKK269 pKa = 9.73IDD271 pKa = 4.47LEE273 pKa = 4.75DD274 pKa = 3.47STSFTVTDD282 pKa = 3.34NAIIVLEE289 pKa = 4.13VIPRR293 pKa = 11.84SSASIHH299 pKa = 5.91SIRR302 pKa = 11.84VAEE305 pKa = 4.29LVTPSSSSIRR315 pKa = 11.84LLHH318 pKa = 6.21TNYY321 pKa = 9.75QQSLMEE327 pKa = 4.37KK328 pKa = 9.41LPNGFTEE335 pKa = 4.09VDD337 pKa = 3.61SYY339 pKa = 10.23PTEE342 pKa = 4.35GSLWSADD349 pKa = 3.96DD350 pKa = 3.72YY351 pKa = 11.91QLYY354 pKa = 9.5QALNQDD360 pKa = 3.07GDD362 pKa = 4.19TVYY365 pKa = 10.24TALIKK370 pKa = 10.6PNNNQFEE377 pKa = 4.42PGNLILLEE385 pKa = 4.2VNLTDD390 pKa = 4.68GSKK393 pKa = 10.34EE394 pKa = 3.95YY395 pKa = 10.95YY396 pKa = 10.22FNSINFKK403 pKa = 10.38FQTIPTDD410 pKa = 3.45QTNWTHH416 pKa = 6.36GGNGSTASPYY426 pKa = 10.84NILDD430 pKa = 3.1TGGRR434 pKa = 11.84VFTWDD439 pKa = 3.37HH440 pKa = 6.51PLDD443 pKa = 3.87EE444 pKa = 5.61FGTEE448 pKa = 4.4LEE450 pKa = 4.08GLNYY454 pKa = 9.92QFEE457 pKa = 4.65LFYY460 pKa = 10.83YY461 pKa = 8.88TDD463 pKa = 3.41NSGTCDD469 pKa = 3.43PHH471 pKa = 8.01VDD473 pKa = 3.8YY474 pKa = 11.27QIGTMQIIDD483 pKa = 3.91FRR485 pKa = 11.84VSDD488 pKa = 4.02LANYY492 pKa = 10.37NDD494 pKa = 3.46ISEE497 pKa = 4.46TEE499 pKa = 3.56IDD501 pKa = 3.86FAGSTTQCLQLDD513 pKa = 3.9IAGSYY518 pKa = 10.22PYY520 pKa = 10.8GDD522 pKa = 3.33NAALKK527 pKa = 10.02IYY529 pKa = 10.37LKK531 pKa = 10.59RR532 pKa = 11.84NSWW535 pKa = 3.2
MM1 pKa = 7.45KK2 pKa = 10.32FKK4 pKa = 11.01LFFIIFTITLLVGCSSSEE22 pKa = 3.74VSTEE26 pKa = 3.38NDD28 pKa = 3.1TVYY31 pKa = 11.22EE32 pKa = 3.91NLTFSGHH39 pKa = 4.3VAIDD43 pKa = 3.67ASSGTSSLEE52 pKa = 3.54ASAYY56 pKa = 10.65ALLSNSNAPSTDD68 pKa = 2.74EE69 pKa = 4.11SNYY72 pKa = 10.14MIVIQNQISNRR83 pKa = 11.84LYY85 pKa = 10.89LSQLDD90 pKa = 3.74ADD92 pKa = 4.42GTFHH96 pKa = 7.62FSDD99 pKa = 3.98NGTDD103 pKa = 3.32AQAIQSVNHH112 pKa = 6.62DD113 pKa = 3.45GGYY116 pKa = 10.54GNHH119 pKa = 5.9FMVMLLQKK127 pKa = 10.92DD128 pKa = 3.75PLEE131 pKa = 4.63VKK133 pKa = 10.43AVAYY137 pKa = 10.55LPGTSEE143 pKa = 3.72EE144 pKa = 5.1GYY146 pKa = 10.76SGFQFSEE153 pKa = 4.37SFDD156 pKa = 3.56QEE158 pKa = 3.99LQFNYY163 pKa = 10.59NSDD166 pKa = 3.96SASMTVSEE174 pKa = 4.21THH176 pKa = 6.61INDD179 pKa = 3.28IAGLQVDD186 pKa = 3.69EE187 pKa = 5.15DD188 pKa = 4.27FVVRR192 pKa = 11.84LVNDD196 pKa = 4.34TPAGATNFGKK206 pKa = 8.6STDD209 pKa = 3.79SQTSTVSDD217 pKa = 3.76INQLDD222 pKa = 4.16PDD224 pKa = 3.81QDD226 pKa = 3.73GQPNIFDD233 pKa = 3.82GMNNGSDD240 pKa = 4.4LDD242 pKa = 4.15NLSADD247 pKa = 3.83NKK249 pKa = 10.2TEE251 pKa = 3.86GATVSDD257 pKa = 5.01SIASSIMFMNLKK269 pKa = 9.73IDD271 pKa = 4.47LEE273 pKa = 4.75DD274 pKa = 3.47STSFTVTDD282 pKa = 3.34NAIIVLEE289 pKa = 4.13VIPRR293 pKa = 11.84SSASIHH299 pKa = 5.91SIRR302 pKa = 11.84VAEE305 pKa = 4.29LVTPSSSSIRR315 pKa = 11.84LLHH318 pKa = 6.21TNYY321 pKa = 9.75QQSLMEE327 pKa = 4.37KK328 pKa = 9.41LPNGFTEE335 pKa = 4.09VDD337 pKa = 3.61SYY339 pKa = 10.23PTEE342 pKa = 4.35GSLWSADD349 pKa = 3.96DD350 pKa = 3.72YY351 pKa = 11.91QLYY354 pKa = 9.5QALNQDD360 pKa = 3.07GDD362 pKa = 4.19TVYY365 pKa = 10.24TALIKK370 pKa = 10.6PNNNQFEE377 pKa = 4.42PGNLILLEE385 pKa = 4.2VNLTDD390 pKa = 4.68GSKK393 pKa = 10.34EE394 pKa = 3.95YY395 pKa = 10.95YY396 pKa = 10.22FNSINFKK403 pKa = 10.38FQTIPTDD410 pKa = 3.45QTNWTHH416 pKa = 6.36GGNGSTASPYY426 pKa = 10.84NILDD430 pKa = 3.1TGGRR434 pKa = 11.84VFTWDD439 pKa = 3.37HH440 pKa = 6.51PLDD443 pKa = 3.87EE444 pKa = 5.61FGTEE448 pKa = 4.4LEE450 pKa = 4.08GLNYY454 pKa = 9.92QFEE457 pKa = 4.65LFYY460 pKa = 10.83YY461 pKa = 8.88TDD463 pKa = 3.41NSGTCDD469 pKa = 3.43PHH471 pKa = 8.01VDD473 pKa = 3.8YY474 pKa = 11.27QIGTMQIIDD483 pKa = 3.91FRR485 pKa = 11.84VSDD488 pKa = 4.02LANYY492 pKa = 10.37NDD494 pKa = 3.46ISEE497 pKa = 4.46TEE499 pKa = 3.56IDD501 pKa = 3.86FAGSTTQCLQLDD513 pKa = 3.9IAGSYY518 pKa = 10.22PYY520 pKa = 10.8GDD522 pKa = 3.33NAALKK527 pKa = 10.02IYY529 pKa = 10.37LKK531 pKa = 10.59RR532 pKa = 11.84NSWW535 pKa = 3.2
Molecular weight: 58.95 kDa
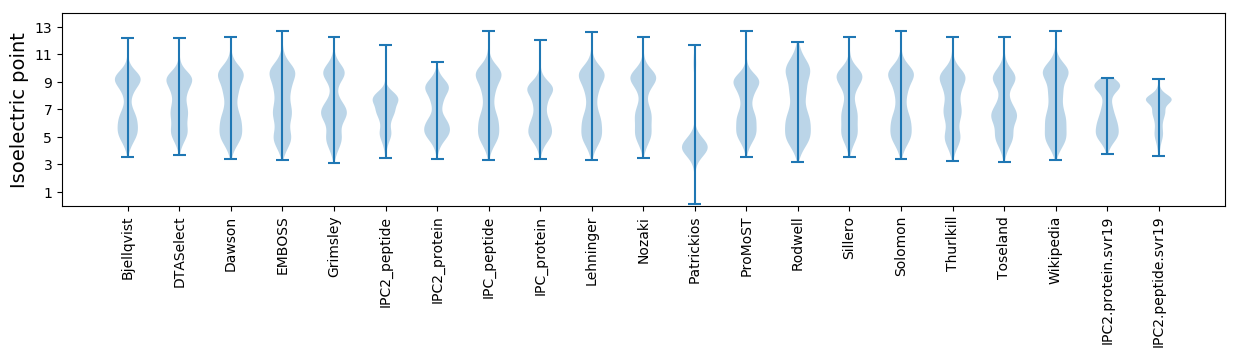
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A328RGJ9|A0A328RGJ9_9BACT Uncharacterized protein OS=Candidatus Marinamargulisbacteria bacterium SCGC AG-414-C22 OX=2184344 GN=DID76_03085 PE=4 SV=1
MM1 pKa = 7.38LRR3 pKa = 11.84SHH5 pKa = 7.07CFASLRR11 pKa = 11.84FALLCIASLRR21 pKa = 11.84IALHH25 pKa = 6.33RR26 pKa = 11.84FAALLCIALLCIALHH41 pKa = 6.45RR42 pKa = 11.84FAALLCIASRR52 pKa = 11.84CFALLRR58 pKa = 11.84FARR61 pKa = 11.84IALFDD66 pKa = 3.99WLSHH70 pKa = 5.55KK71 pKa = 10.71QSFVV75 pKa = 3.02
MM1 pKa = 7.38LRR3 pKa = 11.84SHH5 pKa = 7.07CFASLRR11 pKa = 11.84FALLCIASLRR21 pKa = 11.84IALHH25 pKa = 6.33RR26 pKa = 11.84FAALLCIALLCIALHH41 pKa = 6.45RR42 pKa = 11.84FAALLCIASRR52 pKa = 11.84CFALLRR58 pKa = 11.84FARR61 pKa = 11.84IALFDD66 pKa = 3.99WLSHH70 pKa = 5.55KK71 pKa = 10.71QSFVV75 pKa = 3.02
Molecular weight: 8.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
325261 |
46 |
5061 |
349.7 |
39.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.805 ± 0.087 | 1.301 ± 0.036 |
5.623 ± 0.083 | 5.659 ± 0.096 |
5.215 ± 0.08 | 5.278 ± 0.1 |
2.604 ± 0.045 | 8.32 ± 0.094 |
7.786 ± 0.105 | 10.298 ± 0.108 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.049 ± 0.034 | 5.612 ± 0.082 |
3.502 ± 0.042 | 4.26 ± 0.062 |
3.174 ± 0.058 | 7.072 ± 0.081 |
6.035 ± 0.07 | 5.784 ± 0.079 |
0.818 ± 0.027 | 3.803 ± 0.056 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
