
Halovivax ruber (strain DSM 18193 / JCM 13892 / XH-70)
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Natrialbales; Natrialbaceae; Halovivax; Halovivax ruber
Average proteome isoelectric point is 4.82
Get precalculated fractions of proteins
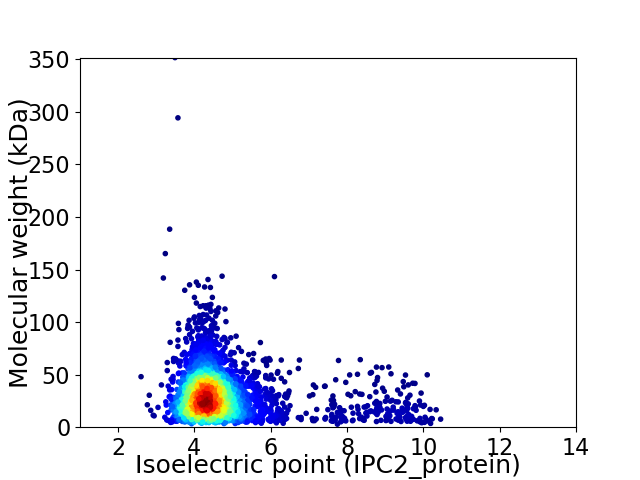
Virtual 2D-PAGE plot for 3099 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|L0II40|L0II40_HALRX Uroporphyrinogen-III synthase OS=Halovivax ruber (strain DSM 18193 / JCM 13892 / XH-70) OX=797302 GN=Halru_3086 PE=4 SV=1
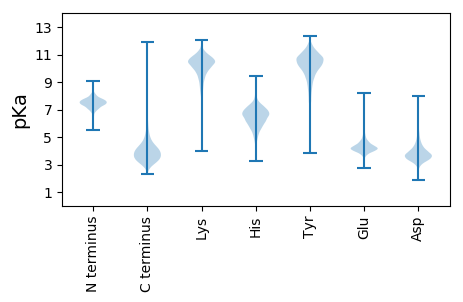
MM1 pKa = 7.46CTRR4 pKa = 11.84DD5 pKa = 3.63SPSSGDD11 pKa = 3.28ARR13 pKa = 11.84PRR15 pKa = 11.84RR16 pKa = 11.84AVLTGAAVVGLSALAGCLDD35 pKa = 4.24GSDD38 pKa = 4.44GDD40 pKa = 5.04ADD42 pKa = 4.01APDD45 pKa = 5.86PITIEE50 pKa = 4.1PDD52 pKa = 3.35RR53 pKa = 11.84ACDD56 pKa = 3.36NCTMRR61 pKa = 11.84IGDD64 pKa = 3.73YY65 pKa = 10.78SGTAGQSFYY74 pKa = 11.18DD75 pKa = 4.0DD76 pKa = 4.17PEE78 pKa = 5.73AVLGPGDD85 pKa = 3.89DD86 pKa = 3.92TDD88 pKa = 5.63RR89 pKa = 11.84PAQFCSARR97 pKa = 11.84CTYY100 pKa = 10.48TFTFDD105 pKa = 3.47NEE107 pKa = 4.11AEE109 pKa = 4.15AEE111 pKa = 4.24PVVSYY116 pKa = 9.21LTDD119 pKa = 3.33YY120 pKa = 11.34SAVDD124 pKa = 3.24WSVDD128 pKa = 3.25TGGAAPTMSRR138 pKa = 11.84HH139 pKa = 5.92LDD141 pKa = 3.15ADD143 pKa = 3.78AFAPAADD150 pKa = 3.82LTLVVDD156 pKa = 4.41SDD158 pKa = 4.32VEE160 pKa = 4.3GAMGRR165 pKa = 11.84SVLGFSNADD174 pKa = 3.5DD175 pKa = 4.42AEE177 pKa = 4.46SFQAEE182 pKa = 4.58HH183 pKa = 6.61GGEE186 pKa = 4.29LYY188 pKa = 10.84DD189 pKa = 5.3HH190 pKa = 7.33EE191 pKa = 5.48DD192 pKa = 3.32VSPTLIQSLMGG203 pKa = 4.01
MM1 pKa = 7.46CTRR4 pKa = 11.84DD5 pKa = 3.63SPSSGDD11 pKa = 3.28ARR13 pKa = 11.84PRR15 pKa = 11.84RR16 pKa = 11.84AVLTGAAVVGLSALAGCLDD35 pKa = 4.24GSDD38 pKa = 4.44GDD40 pKa = 5.04ADD42 pKa = 4.01APDD45 pKa = 5.86PITIEE50 pKa = 4.1PDD52 pKa = 3.35RR53 pKa = 11.84ACDD56 pKa = 3.36NCTMRR61 pKa = 11.84IGDD64 pKa = 3.73YY65 pKa = 10.78SGTAGQSFYY74 pKa = 11.18DD75 pKa = 4.0DD76 pKa = 4.17PEE78 pKa = 5.73AVLGPGDD85 pKa = 3.89DD86 pKa = 3.92TDD88 pKa = 5.63RR89 pKa = 11.84PAQFCSARR97 pKa = 11.84CTYY100 pKa = 10.48TFTFDD105 pKa = 3.47NEE107 pKa = 4.11AEE109 pKa = 4.15AEE111 pKa = 4.24PVVSYY116 pKa = 9.21LTDD119 pKa = 3.33YY120 pKa = 11.34SAVDD124 pKa = 3.24WSVDD128 pKa = 3.25TGGAAPTMSRR138 pKa = 11.84HH139 pKa = 5.92LDD141 pKa = 3.15ADD143 pKa = 3.78AFAPAADD150 pKa = 3.82LTLVVDD156 pKa = 4.41SDD158 pKa = 4.32VEE160 pKa = 4.3GAMGRR165 pKa = 11.84SVLGFSNADD174 pKa = 3.5DD175 pKa = 4.42AEE177 pKa = 4.46SFQAEE182 pKa = 4.58HH183 pKa = 6.61GGEE186 pKa = 4.29LYY188 pKa = 10.84DD189 pKa = 5.3HH190 pKa = 7.33EE191 pKa = 5.48DD192 pKa = 3.32VSPTLIQSLMGG203 pKa = 4.01
Molecular weight: 21.19 kDa
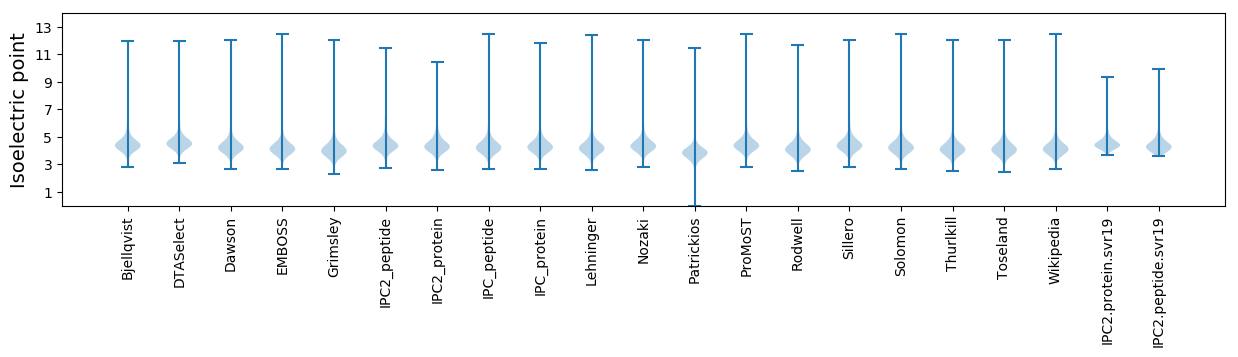
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|L0IG69|L0IG69_HALRX Putative oxidoreductase aryl-alcohol dehydrogenase like protein OS=Halovivax ruber (strain DSM 18193 / JCM 13892 / XH-70) OX=797302 GN=Halru_2644 PE=4 SV=1
MM1 pKa = 7.57TDD3 pKa = 3.01AFVPASALYY12 pKa = 10.29DD13 pKa = 3.46VEE15 pKa = 4.31TRR17 pKa = 11.84STFVHH22 pKa = 6.66PAKK25 pKa = 9.99QASEE29 pKa = 4.02IEE31 pKa = 4.34SPKK34 pKa = 9.94TSGDD38 pKa = 3.65AIEE41 pKa = 5.31ADD43 pKa = 3.88VIQAVDD49 pKa = 3.13ALGYY53 pKa = 10.81VGDD56 pKa = 4.66ATVTWHH62 pKa = 7.11DD63 pKa = 3.77AEE65 pKa = 4.64TTGLLKK71 pKa = 10.6PSTALPFYY79 pKa = 10.05GIVVVMPEE87 pKa = 3.96TPIEE91 pKa = 4.02IEE93 pKa = 3.91ACQVRR98 pKa = 11.84TSNGSRR104 pKa = 11.84STRR107 pKa = 11.84GRR109 pKa = 11.84FYY111 pKa = 10.03VKK113 pKa = 10.07RR114 pKa = 11.84RR115 pKa = 11.84TRR117 pKa = 11.84VAAQRR122 pKa = 11.84SRR124 pKa = 11.84DD125 pKa = 3.63QIRR128 pKa = 11.84GRR130 pKa = 11.84RR131 pKa = 11.84QSRR134 pKa = 11.84RR135 pKa = 11.84TGRR138 pKa = 11.84QSATHH143 pKa = 6.56RR144 pKa = 11.84VRR146 pKa = 11.84NDD148 pKa = 2.83SGPRR152 pKa = 11.84TRR154 pKa = 11.84LHH156 pKa = 5.8GNARR160 pKa = 11.84RR161 pKa = 11.84SS162 pKa = 3.37
MM1 pKa = 7.57TDD3 pKa = 3.01AFVPASALYY12 pKa = 10.29DD13 pKa = 3.46VEE15 pKa = 4.31TRR17 pKa = 11.84STFVHH22 pKa = 6.66PAKK25 pKa = 9.99QASEE29 pKa = 4.02IEE31 pKa = 4.34SPKK34 pKa = 9.94TSGDD38 pKa = 3.65AIEE41 pKa = 5.31ADD43 pKa = 3.88VIQAVDD49 pKa = 3.13ALGYY53 pKa = 10.81VGDD56 pKa = 4.66ATVTWHH62 pKa = 7.11DD63 pKa = 3.77AEE65 pKa = 4.64TTGLLKK71 pKa = 10.6PSTALPFYY79 pKa = 10.05GIVVVMPEE87 pKa = 3.96TPIEE91 pKa = 4.02IEE93 pKa = 3.91ACQVRR98 pKa = 11.84TSNGSRR104 pKa = 11.84STRR107 pKa = 11.84GRR109 pKa = 11.84FYY111 pKa = 10.03VKK113 pKa = 10.07RR114 pKa = 11.84RR115 pKa = 11.84TRR117 pKa = 11.84VAAQRR122 pKa = 11.84SRR124 pKa = 11.84DD125 pKa = 3.63QIRR128 pKa = 11.84GRR130 pKa = 11.84RR131 pKa = 11.84QSRR134 pKa = 11.84RR135 pKa = 11.84TGRR138 pKa = 11.84QSATHH143 pKa = 6.56RR144 pKa = 11.84VRR146 pKa = 11.84NDD148 pKa = 2.83SGPRR152 pKa = 11.84TRR154 pKa = 11.84LHH156 pKa = 5.8GNARR160 pKa = 11.84RR161 pKa = 11.84SS162 pKa = 3.37
Molecular weight: 17.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
914088 |
29 |
3386 |
295.0 |
31.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.092 ± 0.056 | 0.747 ± 0.015 |
8.821 ± 0.056 | 8.23 ± 0.057 |
3.153 ± 0.026 | 8.523 ± 0.043 |
2.046 ± 0.02 | 4.274 ± 0.032 |
1.567 ± 0.025 | 8.72 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.682 ± 0.02 | 2.131 ± 0.022 |
4.808 ± 0.03 | 2.438 ± 0.025 |
6.445 ± 0.044 | 5.858 ± 0.036 |
6.833 ± 0.036 | 8.912 ± 0.044 |
1.118 ± 0.019 | 2.602 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
