
Schistocephalus solidus (Tapeworm)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Spiralia; Lophotrochozoa; Platyhelminthes; Cestoda; Eucestoda; Diphyllobothriidea; Diphyllobothriidae; Schistocephalus
Average proteome isoelectric point is 6.99
Get precalculated fractions of proteins
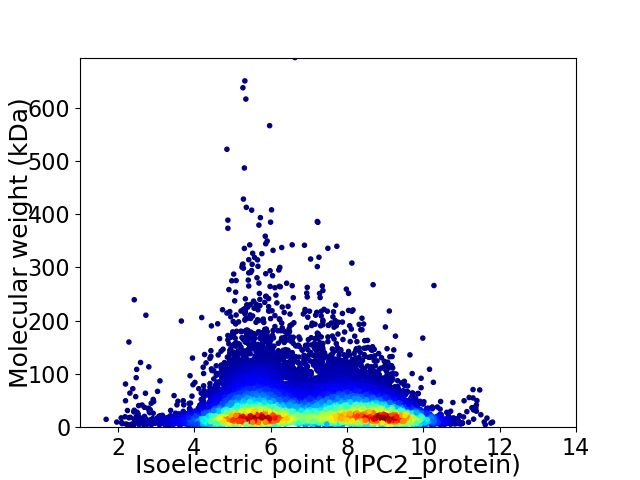
Virtual 2D-PAGE plot for 20172 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A183SMY3|A0A183SMY3_SCHSO Reverse transcriptase domain-containing protein OS=Schistocephalus solidus OX=70667 GN=SSLN_LOCUS5581 PE=4 SV=1
MM1 pKa = 7.45VPRR4 pKa = 11.84CPTCGSLSSEE14 pKa = 4.29YY15 pKa = 10.93DD16 pKa = 4.06DD17 pKa = 6.78DD18 pKa = 7.48DD19 pKa = 7.63DD20 pKa = 7.66DD21 pKa = 7.69DD22 pKa = 7.53DD23 pKa = 7.65DD24 pKa = 7.65DD25 pKa = 7.62DD26 pKa = 7.64DD27 pKa = 7.66DD28 pKa = 7.57DD29 pKa = 7.29DD30 pKa = 7.38DD31 pKa = 6.39EE32 pKa = 8.1DD33 pKa = 6.84DD34 pKa = 6.52DD35 pKa = 7.5DD36 pKa = 7.57DD37 pKa = 7.44DD38 pKa = 7.01DD39 pKa = 7.02DD40 pKa = 6.45GGASSKK46 pKa = 11.11DD47 pKa = 3.64YY48 pKa = 10.91FCWSPPQKK56 pKa = 10.59LGLLWPRR63 pKa = 11.84THH65 pKa = 7.03VYY67 pKa = 10.81APLLANLPQNSCRR80 pKa = 11.84VTYY83 pKa = 9.98GRR85 pKa = 11.84KK86 pKa = 8.21IALYY90 pKa = 9.7LRR92 pKa = 11.84NIFACPDD99 pKa = 3.2SSRR102 pKa = 11.84VGLYY106 pKa = 10.59DD107 pKa = 3.71PLVLVYY113 pKa = 10.95SNSHH117 pKa = 5.26SNKK120 pKa = 10.02SVLKK124 pKa = 10.2CWW126 pKa = 3.42
MM1 pKa = 7.45VPRR4 pKa = 11.84CPTCGSLSSEE14 pKa = 4.29YY15 pKa = 10.93DD16 pKa = 4.06DD17 pKa = 6.78DD18 pKa = 7.48DD19 pKa = 7.63DD20 pKa = 7.66DD21 pKa = 7.69DD22 pKa = 7.53DD23 pKa = 7.65DD24 pKa = 7.65DD25 pKa = 7.62DD26 pKa = 7.64DD27 pKa = 7.66DD28 pKa = 7.57DD29 pKa = 7.29DD30 pKa = 7.38DD31 pKa = 6.39EE32 pKa = 8.1DD33 pKa = 6.84DD34 pKa = 6.52DD35 pKa = 7.5DD36 pKa = 7.57DD37 pKa = 7.44DD38 pKa = 7.01DD39 pKa = 7.02DD40 pKa = 6.45GGASSKK46 pKa = 11.11DD47 pKa = 3.64YY48 pKa = 10.91FCWSPPQKK56 pKa = 10.59LGLLWPRR63 pKa = 11.84THH65 pKa = 7.03VYY67 pKa = 10.81APLLANLPQNSCRR80 pKa = 11.84VTYY83 pKa = 9.98GRR85 pKa = 11.84KK86 pKa = 8.21IALYY90 pKa = 9.7LRR92 pKa = 11.84NIFACPDD99 pKa = 3.2SSRR102 pKa = 11.84VGLYY106 pKa = 10.59DD107 pKa = 3.71PLVLVYY113 pKa = 10.95SNSHH117 pKa = 5.26SNKK120 pKa = 10.02SVLKK124 pKa = 10.2CWW126 pKa = 3.42
Molecular weight: 14.21 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A183SCY2|A0A183SCY2_SCHSO C2H2-type domain-containing protein OS=Schistocephalus solidus OX=70667 GN=SSLN_LOCUS2080 PE=4 SV=1
MM1 pKa = 7.21MMMMMMRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84MTIILMMVVMLMMMVVLVMIMLIMMRR43 pKa = 11.84RR44 pKa = 11.84SMMMMMLIMMMMRR57 pKa = 11.84RR58 pKa = 11.84MTSMMMMMRR67 pKa = 11.84RR68 pKa = 11.84RR69 pKa = 11.84RR70 pKa = 11.84RR71 pKa = 11.84MTVMMRR77 pKa = 11.84RR78 pKa = 11.84RR79 pKa = 11.84RR80 pKa = 11.84MTMIMMMMKK89 pKa = 10.08RR90 pKa = 11.84RR91 pKa = 11.84RR92 pKa = 11.84RR93 pKa = 11.84RR94 pKa = 11.84ITMMMMRR101 pKa = 11.84RR102 pKa = 11.84RR103 pKa = 11.84RR104 pKa = 11.84RR105 pKa = 11.84RR106 pKa = 11.84RR107 pKa = 11.84ITSMMMIMMMIIMMMMMMMMMTAMIMMIMRR137 pKa = 11.84MMMIIIMMMTARR149 pKa = 11.84MIMMMRR155 pKa = 11.84MMKK158 pKa = 10.22RR159 pKa = 11.84RR160 pKa = 11.84MTVMMRR166 pKa = 11.84RR167 pKa = 11.84RR168 pKa = 11.84MTIIMMMMRR177 pKa = 11.84RR178 pKa = 11.84RR179 pKa = 11.84RR180 pKa = 11.84RR181 pKa = 11.84RR182 pKa = 11.84RR183 pKa = 11.84ITMMMRR189 pKa = 11.84RR190 pKa = 11.84RR191 pKa = 11.84RR192 pKa = 11.84MTSMMMMRR200 pKa = 11.84RR201 pKa = 11.84MTMIMMMKK209 pKa = 9.87RR210 pKa = 11.84RR211 pKa = 11.84RR212 pKa = 11.84RR213 pKa = 11.84RR214 pKa = 11.84KK215 pKa = 7.62RR216 pKa = 11.84RR217 pKa = 11.84RR218 pKa = 11.84RR219 pKa = 11.84ITMMMMGVCPPQRR232 pKa = 11.84LWPLKK237 pKa = 8.68VTHH240 pKa = 6.77TEE242 pKa = 3.6TGEE245 pKa = 4.1RR246 pKa = 11.84EE247 pKa = 4.28GG248 pKa = 4.39
MM1 pKa = 7.21MMMMMMRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84MTIILMMVVMLMMMVVLVMIMLIMMRR43 pKa = 11.84RR44 pKa = 11.84SMMMMMLIMMMMRR57 pKa = 11.84RR58 pKa = 11.84MTSMMMMMRR67 pKa = 11.84RR68 pKa = 11.84RR69 pKa = 11.84RR70 pKa = 11.84RR71 pKa = 11.84MTVMMRR77 pKa = 11.84RR78 pKa = 11.84RR79 pKa = 11.84RR80 pKa = 11.84MTMIMMMMKK89 pKa = 10.08RR90 pKa = 11.84RR91 pKa = 11.84RR92 pKa = 11.84RR93 pKa = 11.84RR94 pKa = 11.84ITMMMMRR101 pKa = 11.84RR102 pKa = 11.84RR103 pKa = 11.84RR104 pKa = 11.84RR105 pKa = 11.84RR106 pKa = 11.84RR107 pKa = 11.84ITSMMMIMMMIIMMMMMMMMMTAMIMMIMRR137 pKa = 11.84MMMIIIMMMTARR149 pKa = 11.84MIMMMRR155 pKa = 11.84MMKK158 pKa = 10.22RR159 pKa = 11.84RR160 pKa = 11.84MTVMMRR166 pKa = 11.84RR167 pKa = 11.84RR168 pKa = 11.84MTIIMMMMRR177 pKa = 11.84RR178 pKa = 11.84RR179 pKa = 11.84RR180 pKa = 11.84RR181 pKa = 11.84RR182 pKa = 11.84RR183 pKa = 11.84ITMMMRR189 pKa = 11.84RR190 pKa = 11.84RR191 pKa = 11.84RR192 pKa = 11.84MTSMMMMRR200 pKa = 11.84RR201 pKa = 11.84MTMIMMMKK209 pKa = 9.87RR210 pKa = 11.84RR211 pKa = 11.84RR212 pKa = 11.84RR213 pKa = 11.84RR214 pKa = 11.84KK215 pKa = 7.62RR216 pKa = 11.84RR217 pKa = 11.84RR218 pKa = 11.84RR219 pKa = 11.84ITMMMMGVCPPQRR232 pKa = 11.84LWPLKK237 pKa = 8.68VTHH240 pKa = 6.77TEE242 pKa = 3.6TGEE245 pKa = 4.1RR246 pKa = 11.84EE247 pKa = 4.28GG248 pKa = 4.39
Molecular weight: 32.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6502664 |
29 |
6260 |
322.4 |
35.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.916 ± 0.02 | 2.155 ± 0.011 |
5.28 ± 0.037 | 5.639 ± 0.021 |
3.952 ± 0.017 | 5.73 ± 0.024 |
2.639 ± 0.009 | 4.511 ± 0.015 |
4.314 ± 0.017 | 10.086 ± 0.029 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.19 ± 0.018 | 3.771 ± 0.009 |
6.217 ± 0.024 | 4.115 ± 0.014 |
6.491 ± 0.018 | 8.939 ± 0.028 |
6.245 ± 0.017 | 6.187 ± 0.016 |
1.118 ± 0.007 | 2.483 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
