
Murine polyomavirus (strain A3) (MPyV)
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Alphapolyomavirus; Mus musculus polyomavirus 1
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
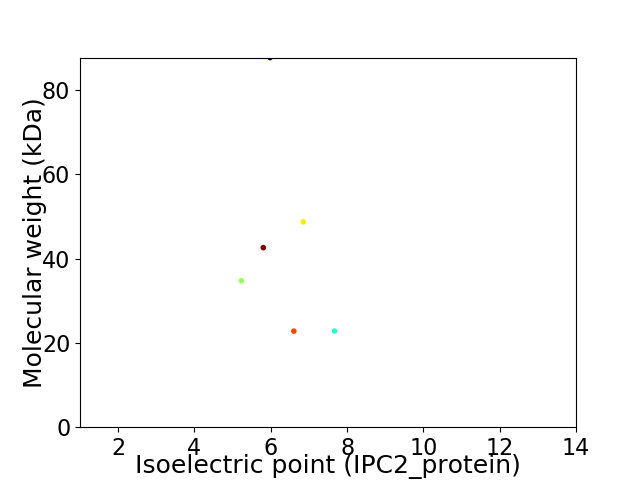
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

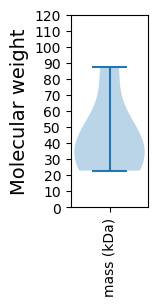
Protein with the lowest isoelectric point:
>sp|P0DOJ5|LT_POVM3 Large T antigen OS=Murine polyomavirus (strain A3) OX=157703 PE=1 SV=1
MM1 pKa = 7.22GAALTILVDD10 pKa = 5.25LIEE13 pKa = 4.62GLAEE17 pKa = 4.01VSTLTGLSAEE27 pKa = 4.95AILSGEE33 pKa = 4.18ALAALDD39 pKa = 4.41GEE41 pKa = 4.62ITALTLEE48 pKa = 4.75GVMSSEE54 pKa = 4.09TALATMGISEE64 pKa = 4.38EE65 pKa = 4.53VYY67 pKa = 10.09GFVSTVPVFVNRR79 pKa = 11.84TAGAIWLMQTVQGASTISLGIQRR102 pKa = 11.84YY103 pKa = 6.9LHH105 pKa = 5.99NEE107 pKa = 3.6EE108 pKa = 4.36VPTVNRR114 pKa = 11.84NMALIPWRR122 pKa = 11.84DD123 pKa = 3.35PALLDD128 pKa = 3.47IYY130 pKa = 10.9FPGVNQFAHH139 pKa = 6.9ALNVVHH145 pKa = 7.45DD146 pKa = 4.14WGHH149 pKa = 5.87GLLHH153 pKa = 6.21SVGRR157 pKa = 11.84YY158 pKa = 4.99VWQMVVQEE166 pKa = 4.3TQHH169 pKa = 6.79RR170 pKa = 11.84LEE172 pKa = 4.31GAVRR176 pKa = 11.84EE177 pKa = 4.19LTVRR181 pKa = 11.84QTHH184 pKa = 5.6TFLDD188 pKa = 4.04GLARR192 pKa = 11.84LLEE195 pKa = 3.97NTRR198 pKa = 11.84WVVSNAPQSAIDD210 pKa = 4.41AINRR214 pKa = 11.84GASSVSSGYY223 pKa = 10.78SSLSDD228 pKa = 3.48YY229 pKa = 10.74YY230 pKa = 10.85RR231 pKa = 11.84QLGLNPPQRR240 pKa = 11.84RR241 pKa = 11.84ALFNRR246 pKa = 11.84IEE248 pKa = 4.06GSMGNGGPTPAAHH261 pKa = 6.53IQDD264 pKa = 3.67EE265 pKa = 4.48SGEE268 pKa = 4.32VIKK271 pKa = 10.28FYY273 pKa = 10.31QAPGGAHH280 pKa = 5.59QRR282 pKa = 11.84VTPDD286 pKa = 2.33WMLPLILGLYY296 pKa = 10.53GDD298 pKa = 4.76ITPTWATVIEE308 pKa = 4.19EE309 pKa = 5.22DD310 pKa = 4.7GPQKK314 pKa = 10.47KK315 pKa = 9.35KK316 pKa = 10.6RR317 pKa = 11.84RR318 pKa = 11.84LL319 pKa = 3.34
MM1 pKa = 7.22GAALTILVDD10 pKa = 5.25LIEE13 pKa = 4.62GLAEE17 pKa = 4.01VSTLTGLSAEE27 pKa = 4.95AILSGEE33 pKa = 4.18ALAALDD39 pKa = 4.41GEE41 pKa = 4.62ITALTLEE48 pKa = 4.75GVMSSEE54 pKa = 4.09TALATMGISEE64 pKa = 4.38EE65 pKa = 4.53VYY67 pKa = 10.09GFVSTVPVFVNRR79 pKa = 11.84TAGAIWLMQTVQGASTISLGIQRR102 pKa = 11.84YY103 pKa = 6.9LHH105 pKa = 5.99NEE107 pKa = 3.6EE108 pKa = 4.36VPTVNRR114 pKa = 11.84NMALIPWRR122 pKa = 11.84DD123 pKa = 3.35PALLDD128 pKa = 3.47IYY130 pKa = 10.9FPGVNQFAHH139 pKa = 6.9ALNVVHH145 pKa = 7.45DD146 pKa = 4.14WGHH149 pKa = 5.87GLLHH153 pKa = 6.21SVGRR157 pKa = 11.84YY158 pKa = 4.99VWQMVVQEE166 pKa = 4.3TQHH169 pKa = 6.79RR170 pKa = 11.84LEE172 pKa = 4.31GAVRR176 pKa = 11.84EE177 pKa = 4.19LTVRR181 pKa = 11.84QTHH184 pKa = 5.6TFLDD188 pKa = 4.04GLARR192 pKa = 11.84LLEE195 pKa = 3.97NTRR198 pKa = 11.84WVVSNAPQSAIDD210 pKa = 4.41AINRR214 pKa = 11.84GASSVSSGYY223 pKa = 10.78SSLSDD228 pKa = 3.48YY229 pKa = 10.74YY230 pKa = 10.85RR231 pKa = 11.84QLGLNPPQRR240 pKa = 11.84RR241 pKa = 11.84ALFNRR246 pKa = 11.84IEE248 pKa = 4.06GSMGNGGPTPAAHH261 pKa = 6.53IQDD264 pKa = 3.67EE265 pKa = 4.48SGEE268 pKa = 4.32VIKK271 pKa = 10.28FYY273 pKa = 10.31QAPGGAHH280 pKa = 5.59QRR282 pKa = 11.84VTPDD286 pKa = 2.33WMLPLILGLYY296 pKa = 10.53GDD298 pKa = 4.76ITPTWATVIEE308 pKa = 4.19EE309 pKa = 5.22DD310 pKa = 4.7GPQKK314 pKa = 10.47KK315 pKa = 9.35KK316 pKa = 10.6RR317 pKa = 11.84RR318 pKa = 11.84LL319 pKa = 3.34
Molecular weight: 34.72 kDa
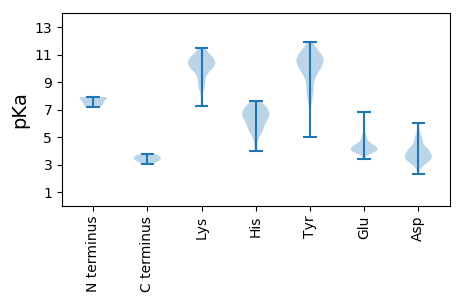
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q84243|Q84243_POVM3 Capsid protein VP1 OS=Murine polyomavirus (strain A3) OX=157703 PE=3 SV=2
MM1 pKa = 7.9DD2 pKa = 4.81RR3 pKa = 11.84VLSRR7 pKa = 11.84ADD9 pKa = 3.34KK10 pKa = 10.73EE11 pKa = 4.2RR12 pKa = 11.84LLEE15 pKa = 4.0LLKK18 pKa = 10.85LPRR21 pKa = 11.84QLWGDD26 pKa = 4.15FGRR29 pKa = 11.84MQQAYY34 pKa = 8.36KK35 pKa = 10.05QQSLLLHH42 pKa = 6.5PDD44 pKa = 2.84KK45 pKa = 11.27GGSHH49 pKa = 7.21ALMQEE54 pKa = 4.36LNSLWGTFKK63 pKa = 10.89TEE65 pKa = 3.95VYY67 pKa = 9.97NLRR70 pKa = 11.84MNLGGTGFQVRR81 pKa = 11.84RR82 pKa = 11.84LHH84 pKa = 6.78ADD86 pKa = 2.98GWNLSTKK93 pKa = 9.7DD94 pKa = 3.51TFGDD98 pKa = 3.34RR99 pKa = 11.84YY100 pKa = 8.3YY101 pKa = 11.23QRR103 pKa = 11.84FCRR106 pKa = 11.84MPLTCLVNVKK116 pKa = 10.13YY117 pKa = 10.71SSCSCILCLLRR128 pKa = 11.84KK129 pKa = 7.28QHH131 pKa = 7.15RR132 pKa = 11.84EE133 pKa = 3.97LKK135 pKa = 9.99DD136 pKa = 3.19KK137 pKa = 11.1CDD139 pKa = 3.54ARR141 pKa = 11.84CLVLGEE147 pKa = 4.41CFCLEE152 pKa = 5.76CYY154 pKa = 7.96MQWFGTPTRR163 pKa = 11.84DD164 pKa = 4.22VLNLYY169 pKa = 10.81ADD171 pKa = 6.04FIASMPIDD179 pKa = 3.67WLDD182 pKa = 3.98LDD184 pKa = 3.83VHH186 pKa = 6.38SVYY189 pKa = 10.77NPRR192 pKa = 11.84LSPP195 pKa = 3.73
MM1 pKa = 7.9DD2 pKa = 4.81RR3 pKa = 11.84VLSRR7 pKa = 11.84ADD9 pKa = 3.34KK10 pKa = 10.73EE11 pKa = 4.2RR12 pKa = 11.84LLEE15 pKa = 4.0LLKK18 pKa = 10.85LPRR21 pKa = 11.84QLWGDD26 pKa = 4.15FGRR29 pKa = 11.84MQQAYY34 pKa = 8.36KK35 pKa = 10.05QQSLLLHH42 pKa = 6.5PDD44 pKa = 2.84KK45 pKa = 11.27GGSHH49 pKa = 7.21ALMQEE54 pKa = 4.36LNSLWGTFKK63 pKa = 10.89TEE65 pKa = 3.95VYY67 pKa = 9.97NLRR70 pKa = 11.84MNLGGTGFQVRR81 pKa = 11.84RR82 pKa = 11.84LHH84 pKa = 6.78ADD86 pKa = 2.98GWNLSTKK93 pKa = 9.7DD94 pKa = 3.51TFGDD98 pKa = 3.34RR99 pKa = 11.84YY100 pKa = 8.3YY101 pKa = 11.23QRR103 pKa = 11.84FCRR106 pKa = 11.84MPLTCLVNVKK116 pKa = 10.13YY117 pKa = 10.71SSCSCILCLLRR128 pKa = 11.84KK129 pKa = 7.28QHH131 pKa = 7.15RR132 pKa = 11.84EE133 pKa = 3.97LKK135 pKa = 9.99DD136 pKa = 3.19KK137 pKa = 11.1CDD139 pKa = 3.54ARR141 pKa = 11.84CLVLGEE147 pKa = 4.41CFCLEE152 pKa = 5.76CYY154 pKa = 7.96MQWFGTPTRR163 pKa = 11.84DD164 pKa = 4.22VLNLYY169 pKa = 10.81ADD171 pKa = 6.04FIASMPIDD179 pKa = 3.67WLDD182 pKa = 3.98LDD184 pKa = 3.83VHH186 pKa = 6.38SVYY189 pKa = 10.77NPRR192 pKa = 11.84LSPP195 pKa = 3.73
Molecular weight: 22.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
2305 |
195 |
782 |
384.2 |
43.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.813 ± 0.713 | 2.299 ± 0.5 |
5.38 ± 0.273 | 5.944 ± 0.357 |
3.514 ± 0.596 | 7.245 ± 0.588 |
2.646 ± 0.369 | 3.341 ± 0.468 |
4.642 ± 0.993 | 11.8 ± 1.087 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.777 ± 0.385 | 3.731 ± 0.433 |
6.334 ± 0.504 | 4.685 ± 0.334 |
6.117 ± 0.868 | 6.855 ± 0.697 |
5.944 ± 0.859 | 5.9 ± 0.857 |
1.735 ± 0.229 | 3.297 ± 0.134 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
