
Candidatus Thiodiazotropha endoloripes
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Gammaproteobacteria incertae sedis; sulfur-oxidizing symbionts; Candidatus Thiodiazotropha
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins
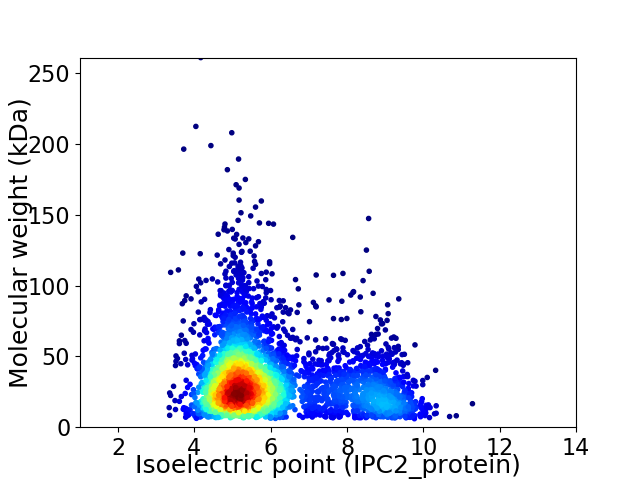
Virtual 2D-PAGE plot for 3951 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1E2UMW2|A0A1E2UMW2_9GAMM SirA family protein OS=Candidatus Thiodiazotropha endoloripes OX=1818881 GN=A3196_04095 PE=3 SV=1
MM1 pKa = 7.1LQKK4 pKa = 10.7KK5 pKa = 7.9SFRR8 pKa = 11.84LLALPASITAAILCSSLLLTGCDD31 pKa = 4.37GDD33 pKa = 5.47DD34 pKa = 4.43GDD36 pKa = 6.29DD37 pKa = 4.87GVDD40 pKa = 3.59GQNGVDD46 pKa = 3.85GTNGTDD52 pKa = 3.49GADD55 pKa = 3.91GITNYY60 pKa = 9.95VPLGLKK66 pKa = 10.17RR67 pKa = 11.84LATAPLDD74 pKa = 3.7AEE76 pKa = 4.42FTGLYY81 pKa = 10.23LNSDD85 pKa = 3.3NTLFLNVQHH94 pKa = 6.97PSSSNTTTDD103 pKa = 2.93AAGKK107 pKa = 10.06VFDD110 pKa = 4.64KK111 pKa = 10.56GTVGVIVGQDD121 pKa = 3.61FSALPEE127 pKa = 4.15NFGALDD133 pKa = 3.93LPVTTAQKK141 pKa = 9.62EE142 pKa = 4.4VVMTAVGSYY151 pKa = 9.07QVLAQQGDD159 pKa = 4.02SLDD162 pKa = 4.7DD163 pKa = 3.91GLAMGDD169 pKa = 3.19IMTADD174 pKa = 3.88GATQIKK180 pKa = 10.18SSNDD184 pKa = 2.96PDD186 pKa = 4.22FNGVVSDD193 pKa = 4.26GNGGFYY199 pKa = 10.61VYY201 pKa = 10.02TNWEE205 pKa = 4.19DD206 pKa = 3.81RR207 pKa = 11.84PGSMSRR213 pKa = 11.84IQVSGLTDD221 pKa = 3.03SGYY224 pKa = 11.52GSITQEE230 pKa = 3.82GMIDD234 pKa = 3.68FSGVGGTWVNCFGTVSPWNTPMSAEE259 pKa = 3.97EE260 pKa = 5.18LYY262 pKa = 10.95FDD264 pKa = 5.48DD265 pKa = 4.42TSDD268 pKa = 3.2WFNPDD273 pKa = 2.67YY274 pKa = 10.93EE275 pKa = 4.58YY276 pKa = 11.12FSNPQSLATYY286 pKa = 10.15LGYY289 pKa = 8.54PTDD292 pKa = 5.54GSGDD296 pKa = 3.11WPNPYY301 pKa = 9.47RR302 pKa = 11.84YY303 pKa = 10.2GYY305 pKa = 9.96IVEE308 pKa = 4.59IGNAADD314 pKa = 3.34AAVANVTVNKK324 pKa = 9.95LEE326 pKa = 4.06TMGRR330 pKa = 11.84FSHH333 pKa = 6.23EE334 pKa = 4.23NSVVMPDD341 pKa = 3.77DD342 pKa = 3.42KK343 pKa = 11.34TVFLSDD349 pKa = 4.01DD350 pKa = 3.46GTGVVFFKK358 pKa = 10.27FVADD362 pKa = 3.67VAGDD366 pKa = 3.62MSAGTLYY373 pKa = 10.81AAQITQAAGVDD384 pKa = 4.12DD385 pKa = 4.41PAEE388 pKa = 4.02AALGIEE394 pKa = 4.42WIEE397 pKa = 3.86LASMGEE403 pKa = 4.11AEE405 pKa = 4.74IEE407 pKa = 3.79AAIASFDD414 pKa = 3.53GTFADD419 pKa = 4.34GNYY422 pKa = 8.23ITDD425 pKa = 3.68EE426 pKa = 4.21QVCDD430 pKa = 3.3WAEE433 pKa = 4.23SKK435 pKa = 10.81SGADD439 pKa = 3.59LSCDD443 pKa = 3.11EE444 pKa = 5.18DD445 pKa = 3.96VTVDD449 pKa = 4.12ANPFSDD455 pKa = 5.13DD456 pKa = 2.86RR457 pKa = 11.84VAYY460 pKa = 10.57LEE462 pKa = 4.0SRR464 pKa = 11.84KK465 pKa = 10.06AAVALGATGEE475 pKa = 4.01FRR477 pKa = 11.84KK478 pKa = 9.65MEE480 pKa = 4.13GVNINYY486 pKa = 9.83NLASTWWNGGAADD499 pKa = 4.17GDD501 pKa = 3.75QAYY504 pKa = 8.6MYY506 pKa = 9.68MAMSSFDD513 pKa = 3.18ATMSDD518 pKa = 4.32DD519 pKa = 3.72EE520 pKa = 5.66GAIQLNGDD528 pKa = 3.54NGKK531 pKa = 10.38CGVVYY536 pKa = 10.59RR537 pKa = 11.84MKK539 pKa = 11.06LMRR542 pKa = 11.84NAAGEE547 pKa = 4.28VDD549 pKa = 4.28VMTMVPAIVGGPYY562 pKa = 10.13YY563 pKa = 10.88ADD565 pKa = 3.35RR566 pKa = 11.84SVNEE570 pKa = 4.25CNVNNISNPDD580 pKa = 3.48NLLIMDD586 pKa = 5.51DD587 pKa = 3.56GRR589 pKa = 11.84VLIGEE594 pKa = 4.12DD595 pKa = 3.21TGNHH599 pKa = 5.36EE600 pKa = 4.66NNVVWVFDD608 pKa = 4.2DD609 pKa = 3.86PAII612 pKa = 4.35
MM1 pKa = 7.1LQKK4 pKa = 10.7KK5 pKa = 7.9SFRR8 pKa = 11.84LLALPASITAAILCSSLLLTGCDD31 pKa = 4.37GDD33 pKa = 5.47DD34 pKa = 4.43GDD36 pKa = 6.29DD37 pKa = 4.87GVDD40 pKa = 3.59GQNGVDD46 pKa = 3.85GTNGTDD52 pKa = 3.49GADD55 pKa = 3.91GITNYY60 pKa = 9.95VPLGLKK66 pKa = 10.17RR67 pKa = 11.84LATAPLDD74 pKa = 3.7AEE76 pKa = 4.42FTGLYY81 pKa = 10.23LNSDD85 pKa = 3.3NTLFLNVQHH94 pKa = 6.97PSSSNTTTDD103 pKa = 2.93AAGKK107 pKa = 10.06VFDD110 pKa = 4.64KK111 pKa = 10.56GTVGVIVGQDD121 pKa = 3.61FSALPEE127 pKa = 4.15NFGALDD133 pKa = 3.93LPVTTAQKK141 pKa = 9.62EE142 pKa = 4.4VVMTAVGSYY151 pKa = 9.07QVLAQQGDD159 pKa = 4.02SLDD162 pKa = 4.7DD163 pKa = 3.91GLAMGDD169 pKa = 3.19IMTADD174 pKa = 3.88GATQIKK180 pKa = 10.18SSNDD184 pKa = 2.96PDD186 pKa = 4.22FNGVVSDD193 pKa = 4.26GNGGFYY199 pKa = 10.61VYY201 pKa = 10.02TNWEE205 pKa = 4.19DD206 pKa = 3.81RR207 pKa = 11.84PGSMSRR213 pKa = 11.84IQVSGLTDD221 pKa = 3.03SGYY224 pKa = 11.52GSITQEE230 pKa = 3.82GMIDD234 pKa = 3.68FSGVGGTWVNCFGTVSPWNTPMSAEE259 pKa = 3.97EE260 pKa = 5.18LYY262 pKa = 10.95FDD264 pKa = 5.48DD265 pKa = 4.42TSDD268 pKa = 3.2WFNPDD273 pKa = 2.67YY274 pKa = 10.93EE275 pKa = 4.58YY276 pKa = 11.12FSNPQSLATYY286 pKa = 10.15LGYY289 pKa = 8.54PTDD292 pKa = 5.54GSGDD296 pKa = 3.11WPNPYY301 pKa = 9.47RR302 pKa = 11.84YY303 pKa = 10.2GYY305 pKa = 9.96IVEE308 pKa = 4.59IGNAADD314 pKa = 3.34AAVANVTVNKK324 pKa = 9.95LEE326 pKa = 4.06TMGRR330 pKa = 11.84FSHH333 pKa = 6.23EE334 pKa = 4.23NSVVMPDD341 pKa = 3.77DD342 pKa = 3.42KK343 pKa = 11.34TVFLSDD349 pKa = 4.01DD350 pKa = 3.46GTGVVFFKK358 pKa = 10.27FVADD362 pKa = 3.67VAGDD366 pKa = 3.62MSAGTLYY373 pKa = 10.81AAQITQAAGVDD384 pKa = 4.12DD385 pKa = 4.41PAEE388 pKa = 4.02AALGIEE394 pKa = 4.42WIEE397 pKa = 3.86LASMGEE403 pKa = 4.11AEE405 pKa = 4.74IEE407 pKa = 3.79AAIASFDD414 pKa = 3.53GTFADD419 pKa = 4.34GNYY422 pKa = 8.23ITDD425 pKa = 3.68EE426 pKa = 4.21QVCDD430 pKa = 3.3WAEE433 pKa = 4.23SKK435 pKa = 10.81SGADD439 pKa = 3.59LSCDD443 pKa = 3.11EE444 pKa = 5.18DD445 pKa = 3.96VTVDD449 pKa = 4.12ANPFSDD455 pKa = 5.13DD456 pKa = 2.86RR457 pKa = 11.84VAYY460 pKa = 10.57LEE462 pKa = 4.0SRR464 pKa = 11.84KK465 pKa = 10.06AAVALGATGEE475 pKa = 4.01FRR477 pKa = 11.84KK478 pKa = 9.65MEE480 pKa = 4.13GVNINYY486 pKa = 9.83NLASTWWNGGAADD499 pKa = 4.17GDD501 pKa = 3.75QAYY504 pKa = 8.6MYY506 pKa = 9.68MAMSSFDD513 pKa = 3.18ATMSDD518 pKa = 4.32DD519 pKa = 3.72EE520 pKa = 5.66GAIQLNGDD528 pKa = 3.54NGKK531 pKa = 10.38CGVVYY536 pKa = 10.59RR537 pKa = 11.84MKK539 pKa = 11.06LMRR542 pKa = 11.84NAAGEE547 pKa = 4.28VDD549 pKa = 4.28VMTMVPAIVGGPYY562 pKa = 10.13YY563 pKa = 10.88ADD565 pKa = 3.35RR566 pKa = 11.84SVNEE570 pKa = 4.25CNVNNISNPDD580 pKa = 3.48NLLIMDD586 pKa = 5.51DD587 pKa = 3.56GRR589 pKa = 11.84VLIGEE594 pKa = 4.12DD595 pKa = 3.21TGNHH599 pKa = 5.36EE600 pKa = 4.66NNVVWVFDD608 pKa = 4.2DD609 pKa = 3.86PAII612 pKa = 4.35
Molecular weight: 65.01 kDa
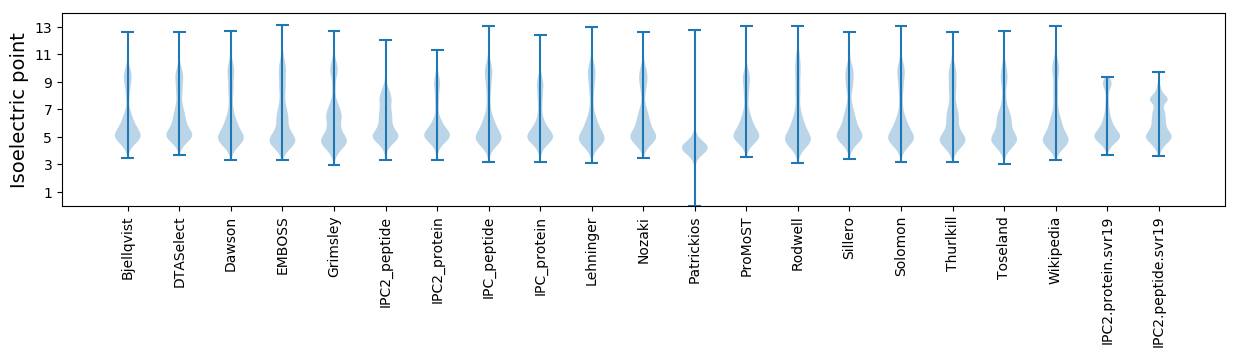
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1E2UMA6|A0A1E2UMA6_9GAMM DNA-binding response regulator OS=Candidatus Thiodiazotropha endoloripes OX=1818881 GN=A3196_03395 PE=4 SV=1
MM1 pKa = 7.38YY2 pKa = 8.15MAKK5 pKa = 10.13KK6 pKa = 10.0KK7 pKa = 9.87AAKK10 pKa = 9.88KK11 pKa = 9.94KK12 pKa = 8.93ATAKK16 pKa = 10.47RR17 pKa = 11.84ATTTKK22 pKa = 10.13KK23 pKa = 10.04VAVKK27 pKa = 10.25KK28 pKa = 10.36RR29 pKa = 11.84ATKK32 pKa = 10.51KK33 pKa = 10.26KK34 pKa = 8.43VATKK38 pKa = 10.53KK39 pKa = 10.29RR40 pKa = 11.84VATKK44 pKa = 10.57KK45 pKa = 10.23KK46 pKa = 8.21ATKK49 pKa = 10.23KK50 pKa = 10.18KK51 pKa = 9.16VAAKK55 pKa = 10.11KK56 pKa = 9.73RR57 pKa = 11.84ATKK60 pKa = 10.33KK61 pKa = 10.33RR62 pKa = 11.84VVKK65 pKa = 10.75KK66 pKa = 10.33KK67 pKa = 8.67ATKK70 pKa = 10.36KK71 pKa = 10.31KK72 pKa = 8.82VAKK75 pKa = 10.34KK76 pKa = 10.05KK77 pKa = 8.6ATKK80 pKa = 10.36KK81 pKa = 10.21KK82 pKa = 8.61VAKK85 pKa = 10.37KK86 pKa = 9.94KK87 pKa = 8.69VAKK90 pKa = 10.41KK91 pKa = 10.0KK92 pKa = 8.69VAKK95 pKa = 10.41KK96 pKa = 10.17KK97 pKa = 8.76VAKK100 pKa = 10.36KK101 pKa = 10.09KK102 pKa = 8.6ATKK105 pKa = 10.36KK106 pKa = 10.21KK107 pKa = 8.61VAKK110 pKa = 10.37KK111 pKa = 10.13KK112 pKa = 8.76VAKK115 pKa = 10.36KK116 pKa = 10.28KK117 pKa = 8.77ATKK120 pKa = 10.18KK121 pKa = 9.97KK122 pKa = 9.18AAKK125 pKa = 9.94KK126 pKa = 9.89KK127 pKa = 9.0VAAKK131 pKa = 9.84KK132 pKa = 10.2RR133 pKa = 11.84PARR136 pKa = 11.84PRR138 pKa = 11.84KK139 pKa = 7.44VTPRR143 pKa = 11.84KK144 pKa = 9.86KK145 pKa = 10.11KK146 pKa = 9.98AASS149 pKa = 3.22
MM1 pKa = 7.38YY2 pKa = 8.15MAKK5 pKa = 10.13KK6 pKa = 10.0KK7 pKa = 9.87AAKK10 pKa = 9.88KK11 pKa = 9.94KK12 pKa = 8.93ATAKK16 pKa = 10.47RR17 pKa = 11.84ATTTKK22 pKa = 10.13KK23 pKa = 10.04VAVKK27 pKa = 10.25KK28 pKa = 10.36RR29 pKa = 11.84ATKK32 pKa = 10.51KK33 pKa = 10.26KK34 pKa = 8.43VATKK38 pKa = 10.53KK39 pKa = 10.29RR40 pKa = 11.84VATKK44 pKa = 10.57KK45 pKa = 10.23KK46 pKa = 8.21ATKK49 pKa = 10.23KK50 pKa = 10.18KK51 pKa = 9.16VAAKK55 pKa = 10.11KK56 pKa = 9.73RR57 pKa = 11.84ATKK60 pKa = 10.33KK61 pKa = 10.33RR62 pKa = 11.84VVKK65 pKa = 10.75KK66 pKa = 10.33KK67 pKa = 8.67ATKK70 pKa = 10.36KK71 pKa = 10.31KK72 pKa = 8.82VAKK75 pKa = 10.34KK76 pKa = 10.05KK77 pKa = 8.6ATKK80 pKa = 10.36KK81 pKa = 10.21KK82 pKa = 8.61VAKK85 pKa = 10.37KK86 pKa = 9.94KK87 pKa = 8.69VAKK90 pKa = 10.41KK91 pKa = 10.0KK92 pKa = 8.69VAKK95 pKa = 10.41KK96 pKa = 10.17KK97 pKa = 8.76VAKK100 pKa = 10.36KK101 pKa = 10.09KK102 pKa = 8.6ATKK105 pKa = 10.36KK106 pKa = 10.21KK107 pKa = 8.61VAKK110 pKa = 10.37KK111 pKa = 10.13KK112 pKa = 8.76VAKK115 pKa = 10.36KK116 pKa = 10.28KK117 pKa = 8.77ATKK120 pKa = 10.18KK121 pKa = 9.97KK122 pKa = 9.18AAKK125 pKa = 9.94KK126 pKa = 9.89KK127 pKa = 9.0VAAKK131 pKa = 9.84KK132 pKa = 10.2RR133 pKa = 11.84PARR136 pKa = 11.84PRR138 pKa = 11.84KK139 pKa = 7.44VTPRR143 pKa = 11.84KK144 pKa = 9.86KK145 pKa = 10.11KK146 pKa = 9.98AASS149 pKa = 3.22
Molecular weight: 16.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1269643 |
51 |
2344 |
321.3 |
35.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.523 ± 0.039 | 0.988 ± 0.018 |
5.807 ± 0.034 | 6.664 ± 0.042 |
3.711 ± 0.022 | 7.324 ± 0.04 |
2.338 ± 0.023 | 6.067 ± 0.033 |
4.326 ± 0.032 | 10.963 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.62 ± 0.019 | 3.634 ± 0.03 |
4.392 ± 0.027 | 4.664 ± 0.035 |
5.712 ± 0.034 | 6.431 ± 0.028 |
5.043 ± 0.035 | 6.648 ± 0.033 |
1.285 ± 0.016 | 2.858 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
