
Pelagibacterium halotolerans (strain DSM 22347 / JCM 15775 / CGMCC 1.7692 / B2)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Devosiaceae; Pelagibacterium; Pelagibacterium halotolerans
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
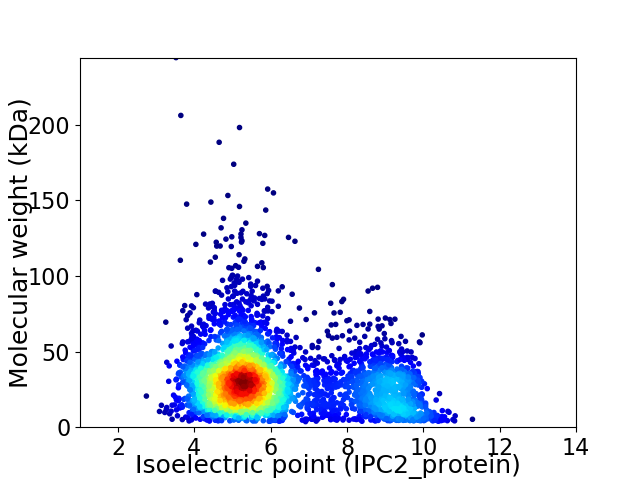
Virtual 2D-PAGE plot for 3875 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G4RF18|G4RF18_PELHB Uncharacterized protein OS=Pelagibacterium halotolerans (strain DSM 22347 / JCM 15775 / CGMCC 1.7692 / B2) OX=1082931 GN=KKY_2956 PE=4 SV=1
MM1 pKa = 7.64LLLSSSTEE9 pKa = 3.88GFEE12 pKa = 4.4MKK14 pKa = 10.29RR15 pKa = 11.84LLLIGTAAYY24 pKa = 9.93AVLGSAAMAAEE35 pKa = 4.61PVTLWFWGAPPNLQDD50 pKa = 3.51AFEE53 pKa = 4.27EE54 pKa = 4.56VLVGPFNASQDD65 pKa = 3.61EE66 pKa = 4.52YY67 pKa = 10.92EE68 pKa = 4.23LQIEE72 pKa = 4.54FLQDD76 pKa = 2.36VDD78 pKa = 3.66NDD80 pKa = 3.11VRR82 pKa = 11.84TAVLAGEE89 pKa = 4.91GPDD92 pKa = 3.61LVYY95 pKa = 10.76TSGPSYY101 pKa = 10.43IAPLARR107 pKa = 11.84AGAIEE112 pKa = 4.48PLDD115 pKa = 4.29AYY117 pKa = 10.63AEE119 pKa = 4.2QYY121 pKa = 9.72GWHH124 pKa = 7.11DD125 pKa = 3.69RR126 pKa = 11.84LLEE129 pKa = 4.14PVLDD133 pKa = 3.53TCYY136 pKa = 10.7QLDD139 pKa = 4.17HH140 pKa = 7.15LYY142 pKa = 10.98CMPPALISDD151 pKa = 3.48GMFYY155 pKa = 11.03NRR157 pKa = 11.84ALLEE161 pKa = 4.22EE162 pKa = 4.92KK163 pKa = 10.09GWEE166 pKa = 4.29VPTTLAEE173 pKa = 4.22VEE175 pKa = 4.33QVMDD179 pKa = 3.98AAIADD184 pKa = 3.83GLYY187 pKa = 10.95ASVTGNKK194 pKa = 8.25GWQPVNEE201 pKa = 4.17NYY203 pKa = 11.05ASIFINNVVGPARR216 pKa = 11.84FYY218 pKa = 10.97EE219 pKa = 4.06ILSTGEE225 pKa = 4.1GWDD228 pKa = 3.39SAEE231 pKa = 3.92MIKK234 pKa = 10.51AIEE237 pKa = 4.02EE238 pKa = 3.87SARR241 pKa = 11.84WFKK244 pKa = 11.04AGYY247 pKa = 9.86LGGSDD252 pKa = 3.74YY253 pKa = 11.13FSLNFDD259 pKa = 3.52EE260 pKa = 5.85SISLVSQKK268 pKa = 10.37RR269 pKa = 11.84SPFFFAPSIGFQWATNYY286 pKa = 7.5FTGDD290 pKa = 3.26AAGDD294 pKa = 3.52FAFAPIPQMDD304 pKa = 3.96EE305 pKa = 3.9SLPYY309 pKa = 9.85PIYY312 pKa = 10.64DD313 pKa = 3.55LGVAFTLSINANSDD327 pKa = 3.58VKK329 pKa = 11.1DD330 pKa = 3.58GAAAVLDD337 pKa = 5.07LIFSPEE343 pKa = 3.98FASDD347 pKa = 3.71MADD350 pKa = 2.79VWPGYY355 pKa = 9.3WGIPLRR361 pKa = 11.84EE362 pKa = 4.23FPTNPDD368 pKa = 3.0ATGLTASFLDD378 pKa = 3.81AMADD382 pKa = 3.14MTAAVDD388 pKa = 3.26AGTFGFKK395 pKa = 9.87IGTFFPPATSQVMFEE410 pKa = 5.11DD411 pKa = 4.69IEE413 pKa = 4.95SVWLDD418 pKa = 3.08RR419 pKa = 11.84MTAQDD424 pKa = 4.21MLAKK428 pKa = 10.17AASAYY433 pKa = 9.9ADD435 pKa = 3.56EE436 pKa = 4.65MAQGLTQDD444 pKa = 4.54LPQPSMM450 pKa = 3.8
MM1 pKa = 7.64LLLSSSTEE9 pKa = 3.88GFEE12 pKa = 4.4MKK14 pKa = 10.29RR15 pKa = 11.84LLLIGTAAYY24 pKa = 9.93AVLGSAAMAAEE35 pKa = 4.61PVTLWFWGAPPNLQDD50 pKa = 3.51AFEE53 pKa = 4.27EE54 pKa = 4.56VLVGPFNASQDD65 pKa = 3.61EE66 pKa = 4.52YY67 pKa = 10.92EE68 pKa = 4.23LQIEE72 pKa = 4.54FLQDD76 pKa = 2.36VDD78 pKa = 3.66NDD80 pKa = 3.11VRR82 pKa = 11.84TAVLAGEE89 pKa = 4.91GPDD92 pKa = 3.61LVYY95 pKa = 10.76TSGPSYY101 pKa = 10.43IAPLARR107 pKa = 11.84AGAIEE112 pKa = 4.48PLDD115 pKa = 4.29AYY117 pKa = 10.63AEE119 pKa = 4.2QYY121 pKa = 9.72GWHH124 pKa = 7.11DD125 pKa = 3.69RR126 pKa = 11.84LLEE129 pKa = 4.14PVLDD133 pKa = 3.53TCYY136 pKa = 10.7QLDD139 pKa = 4.17HH140 pKa = 7.15LYY142 pKa = 10.98CMPPALISDD151 pKa = 3.48GMFYY155 pKa = 11.03NRR157 pKa = 11.84ALLEE161 pKa = 4.22EE162 pKa = 4.92KK163 pKa = 10.09GWEE166 pKa = 4.29VPTTLAEE173 pKa = 4.22VEE175 pKa = 4.33QVMDD179 pKa = 3.98AAIADD184 pKa = 3.83GLYY187 pKa = 10.95ASVTGNKK194 pKa = 8.25GWQPVNEE201 pKa = 4.17NYY203 pKa = 11.05ASIFINNVVGPARR216 pKa = 11.84FYY218 pKa = 10.97EE219 pKa = 4.06ILSTGEE225 pKa = 4.1GWDD228 pKa = 3.39SAEE231 pKa = 3.92MIKK234 pKa = 10.51AIEE237 pKa = 4.02EE238 pKa = 3.87SARR241 pKa = 11.84WFKK244 pKa = 11.04AGYY247 pKa = 9.86LGGSDD252 pKa = 3.74YY253 pKa = 11.13FSLNFDD259 pKa = 3.52EE260 pKa = 5.85SISLVSQKK268 pKa = 10.37RR269 pKa = 11.84SPFFFAPSIGFQWATNYY286 pKa = 7.5FTGDD290 pKa = 3.26AAGDD294 pKa = 3.52FAFAPIPQMDD304 pKa = 3.96EE305 pKa = 3.9SLPYY309 pKa = 9.85PIYY312 pKa = 10.64DD313 pKa = 3.55LGVAFTLSINANSDD327 pKa = 3.58VKK329 pKa = 11.1DD330 pKa = 3.58GAAAVLDD337 pKa = 5.07LIFSPEE343 pKa = 3.98FASDD347 pKa = 3.71MADD350 pKa = 2.79VWPGYY355 pKa = 9.3WGIPLRR361 pKa = 11.84EE362 pKa = 4.23FPTNPDD368 pKa = 3.0ATGLTASFLDD378 pKa = 3.81AMADD382 pKa = 3.14MTAAVDD388 pKa = 3.26AGTFGFKK395 pKa = 9.87IGTFFPPATSQVMFEE410 pKa = 5.11DD411 pKa = 4.69IEE413 pKa = 4.95SVWLDD418 pKa = 3.08RR419 pKa = 11.84MTAQDD424 pKa = 4.21MLAKK428 pKa = 10.17AASAYY433 pKa = 9.9ADD435 pKa = 3.56EE436 pKa = 4.65MAQGLTQDD444 pKa = 4.54LPQPSMM450 pKa = 3.8
Molecular weight: 49.14 kDa
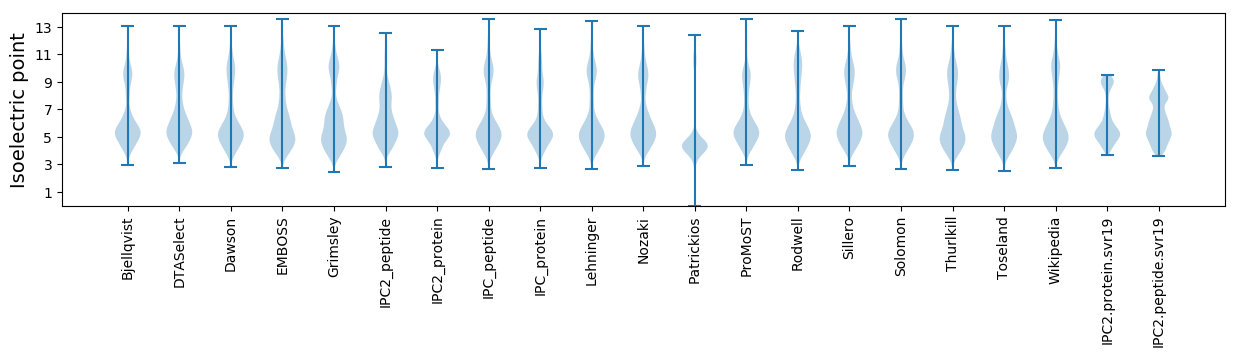
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G4RGH2|G4RGH2_PELHB Uncharacterized protein OS=Pelagibacterium halotolerans (strain DSM 22347 / JCM 15775 / CGMCC 1.7692 / B2) OX=1082931 GN=KKY_101 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.36NGRR28 pKa = 11.84KK29 pKa = 9.11IINRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AIGRR39 pKa = 11.84KK40 pKa = 9.12RR41 pKa = 11.84LSAA44 pKa = 3.98
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.36NGRR28 pKa = 11.84KK29 pKa = 9.11IINRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AIGRR39 pKa = 11.84KK40 pKa = 9.12RR41 pKa = 11.84LSAA44 pKa = 3.98
Molecular weight: 5.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1168210 |
37 |
2379 |
301.5 |
32.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.203 ± 0.055 | 0.706 ± 0.012 |
5.879 ± 0.037 | 5.971 ± 0.041 |
3.914 ± 0.026 | 8.659 ± 0.044 |
1.941 ± 0.019 | 5.79 ± 0.03 |
2.908 ± 0.036 | 10.053 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.65 ± 0.02 | 2.707 ± 0.023 |
4.993 ± 0.029 | 3.101 ± 0.026 |
6.517 ± 0.036 | 5.513 ± 0.03 |
5.481 ± 0.031 | 7.447 ± 0.035 |
1.29 ± 0.018 | 2.277 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
