
Methanoculleus bourgensis
Taxonomy: cellular organisms; Archaea;
Average proteome isoelectric point is 6.38
Get precalculated fractions of proteins
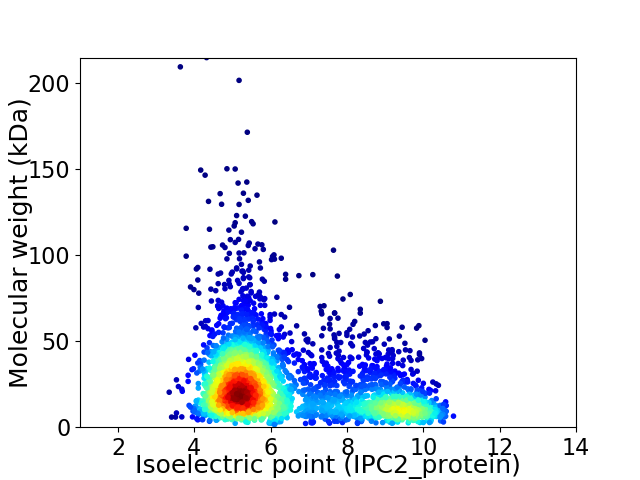
Virtual 2D-PAGE plot for 3446 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0X3BMZ6|A0A0X3BMZ6_9EURY Uncharacterized protein OS=Methanoculleus bourgensis OX=83986 GN=MMAB1_2148 PE=4 SV=1
MM1 pKa = 7.63LSLMLISVVSASYY14 pKa = 11.2PMFHH18 pKa = 7.28YY19 pKa = 7.87DD20 pKa = 3.43TQRR23 pKa = 11.84TGYY26 pKa = 9.55IPQDD30 pKa = 3.56GPQTNATLWVAEE42 pKa = 4.11TAEE45 pKa = 4.28YY46 pKa = 11.04ADD48 pKa = 4.53GSPAVYY54 pKa = 9.75NGKK57 pKa = 9.39VFVPTWPDD65 pKa = 2.48MDD67 pKa = 4.6FADD70 pKa = 4.91NDD72 pKa = 3.74PMGLVCYY79 pKa = 10.13DD80 pKa = 3.34AATGTEE86 pKa = 3.99LWTNEE91 pKa = 4.03LGGTSVGSVSGVAVADD107 pKa = 3.56GRR109 pKa = 11.84VYY111 pKa = 11.04LGGTDD116 pKa = 3.22GRR118 pKa = 11.84LYY120 pKa = 10.96CIDD123 pKa = 4.11EE124 pKa = 4.38EE125 pKa = 4.33TGEE128 pKa = 4.32MFWTSDD134 pKa = 3.39QIDD137 pKa = 3.46ATGYY141 pKa = 10.15FGLSSSPLIYY151 pKa = 10.57EE152 pKa = 3.83GTVYY156 pKa = 10.74VLSASDD162 pKa = 3.88GVLHH166 pKa = 7.2AFTPEE171 pKa = 4.66GIEE174 pKa = 3.9SWSFPTGGAVGYY186 pKa = 7.29FTSPAAANGEE196 pKa = 4.01IFVAGNEE203 pKa = 4.22SDD205 pKa = 4.42LSCIDD210 pKa = 3.3ITTHH214 pKa = 5.01TATWSVALPTAVKK227 pKa = 8.76STPAIGDD234 pKa = 3.33GKK236 pKa = 11.14VYY238 pKa = 9.58VTTAEE243 pKa = 3.82RR244 pKa = 11.84LYY246 pKa = 11.11ALSASTGAEE255 pKa = 3.7VWNASIGGTSSTPAVAGEE273 pKa = 4.1TVIAGSSDD281 pKa = 3.2GLHH284 pKa = 6.92AYY286 pKa = 9.6DD287 pKa = 5.18AGTGTPLWNFPSARR301 pKa = 11.84VDD303 pKa = 3.22VSPIIAGNLVYY314 pKa = 10.47AATNEE319 pKa = 4.34EE320 pKa = 4.25IGTVYY325 pKa = 10.74AVDD328 pKa = 3.26TGTGEE333 pKa = 4.42EE334 pKa = 4.29VWSYY338 pKa = 10.74TIEE341 pKa = 4.24APGDD345 pKa = 3.37GTFAAFFASSPAVSDD360 pKa = 3.53GVLYY364 pKa = 10.46IGVEE368 pKa = 4.05NNRR371 pKa = 11.84LYY373 pKa = 11.17AFGEE377 pKa = 4.74GSVSPTPTPTPTPTPAPGSWNGTVILAKK405 pKa = 9.54EE406 pKa = 4.44TFTFTPSNNASATYY420 pKa = 8.79TVNRR424 pKa = 11.84TTDD427 pKa = 3.49LGALTLAAQAGGFTINASDD446 pKa = 3.49AWYY449 pKa = 10.44ADD451 pKa = 3.35YY452 pKa = 11.19GSFMLEE458 pKa = 4.46DD459 pKa = 3.19IAGIANEE466 pKa = 4.9DD467 pKa = 3.25WTQEE471 pKa = 3.97NARR474 pKa = 11.84SWSIFINGAMAPAGLGANTLADD496 pKa = 3.82GDD498 pKa = 4.0RR499 pKa = 11.84LAFYY503 pKa = 9.66YY504 pKa = 10.65CPSDD508 pKa = 3.68PDD510 pKa = 3.68TYY512 pKa = 11.71APLIDD517 pKa = 3.44QAGYY521 pKa = 10.83VVTIDD526 pKa = 3.29VNVRR530 pKa = 11.84DD531 pKa = 4.58FTWEE535 pKa = 4.03GAVSLTDD542 pKa = 3.52GQTFTITPFNNEE554 pKa = 4.0SAIHH558 pKa = 5.12TFNRR562 pKa = 11.84TSALGALDD570 pKa = 3.82AAATAGGFNYY580 pKa = 9.48TVQEE584 pKa = 4.46TTWGPFLYY592 pKa = 10.46SIGGIAYY599 pKa = 10.29NEE601 pKa = 4.26TSWDD605 pKa = 3.08SWLYY609 pKa = 10.21SVNGIDD615 pKa = 4.45ASVGAADD622 pKa = 3.81YY623 pKa = 11.08QLTDD627 pKa = 3.82GDD629 pKa = 5.05VITYY633 pKa = 8.76WYY635 pKa = 9.08GAWGSTPDD643 pKa = 3.33TAGAVVDD650 pKa = 3.67ITVSIPATPAPTPTSGGGGGGGDD673 pKa = 3.87SPPSRR678 pKa = 11.84ITVTLQPGTFTITAEE693 pKa = 4.09NSGKK697 pKa = 9.38NHH699 pKa = 5.02TVSRR703 pKa = 11.84QTALGALDD711 pKa = 3.68ATGIAYY717 pKa = 8.52TIDD720 pKa = 3.16DD721 pKa = 4.42SYY723 pKa = 9.25YY724 pKa = 10.11QEE726 pKa = 4.39YY727 pKa = 11.0GSLFLSSIRR736 pKa = 11.84GRR738 pKa = 11.84VSEE741 pKa = 4.26GTRR744 pKa = 11.84GWMYY748 pKa = 10.47RR749 pKa = 11.84VNGGSPAVGANAYY762 pKa = 8.15PVTSGDD768 pKa = 3.48DD769 pKa = 3.82VIFFWSEE776 pKa = 4.09SMSSTPATSPDD787 pKa = 3.61VISIRR792 pKa = 11.84VVIPASSGSSGSDD805 pKa = 2.92SSGGGGSGSVPSSTTQEE822 pKa = 4.05QPNSVDD828 pKa = 3.09NSSASFLFGLPDD840 pKa = 3.61GAVIEE845 pKa = 4.3LGEE848 pKa = 4.05WGQTFSINTGPASAGEE864 pKa = 4.15EE865 pKa = 4.09VTISGNTFTINQSGIVLTIVARR887 pKa = 11.84NIDD890 pKa = 3.95EE891 pKa = 4.44KK892 pKa = 11.44DD893 pKa = 3.55GVASGLIEE901 pKa = 4.52SVTAAIDD908 pKa = 4.47LISGEE913 pKa = 4.03IEE915 pKa = 4.59GIGVVAASLDD925 pKa = 3.92LNLTGIPAADD935 pKa = 3.56GRR937 pKa = 11.84LDD939 pKa = 3.21ITFNATPDD947 pKa = 3.43ATAGNAFTLAATEE960 pKa = 3.88NDD962 pKa = 3.59EE963 pKa = 5.33EE964 pKa = 4.68IDD966 pKa = 3.5ALAYY970 pKa = 9.62TMTVTRR976 pKa = 11.84TNLEE980 pKa = 3.83NGEE983 pKa = 4.95DD984 pKa = 3.14ITGAVIRR991 pKa = 11.84MTINPEE997 pKa = 3.39WVEE1000 pKa = 3.72EE1001 pKa = 4.15HH1002 pKa = 6.59GGVDD1006 pKa = 3.24AVRR1009 pKa = 11.84IARR1012 pKa = 11.84SAEE1015 pKa = 3.92DD1016 pKa = 3.52STHH1019 pKa = 6.76EE1020 pKa = 4.36ILDD1023 pKa = 3.6TRR1025 pKa = 11.84LAGTDD1030 pKa = 3.39EE1031 pKa = 4.22NGNLIFEE1038 pKa = 4.54AVSPGGLSVFGLLTVRR1054 pKa = 11.84PASEE1058 pKa = 4.08MQEE1061 pKa = 4.41TPAVTATDD1069 pKa = 4.22PVSSTPVAAGTPEE1082 pKa = 4.19GAPPSLSSPFIGVGVGAALLIGAAYY1107 pKa = 10.23LIIRR1111 pKa = 11.84WRR1113 pKa = 11.84RR1114 pKa = 11.84EE1115 pKa = 3.43RR1116 pKa = 3.89
MM1 pKa = 7.63LSLMLISVVSASYY14 pKa = 11.2PMFHH18 pKa = 7.28YY19 pKa = 7.87DD20 pKa = 3.43TQRR23 pKa = 11.84TGYY26 pKa = 9.55IPQDD30 pKa = 3.56GPQTNATLWVAEE42 pKa = 4.11TAEE45 pKa = 4.28YY46 pKa = 11.04ADD48 pKa = 4.53GSPAVYY54 pKa = 9.75NGKK57 pKa = 9.39VFVPTWPDD65 pKa = 2.48MDD67 pKa = 4.6FADD70 pKa = 4.91NDD72 pKa = 3.74PMGLVCYY79 pKa = 10.13DD80 pKa = 3.34AATGTEE86 pKa = 3.99LWTNEE91 pKa = 4.03LGGTSVGSVSGVAVADD107 pKa = 3.56GRR109 pKa = 11.84VYY111 pKa = 11.04LGGTDD116 pKa = 3.22GRR118 pKa = 11.84LYY120 pKa = 10.96CIDD123 pKa = 4.11EE124 pKa = 4.38EE125 pKa = 4.33TGEE128 pKa = 4.32MFWTSDD134 pKa = 3.39QIDD137 pKa = 3.46ATGYY141 pKa = 10.15FGLSSSPLIYY151 pKa = 10.57EE152 pKa = 3.83GTVYY156 pKa = 10.74VLSASDD162 pKa = 3.88GVLHH166 pKa = 7.2AFTPEE171 pKa = 4.66GIEE174 pKa = 3.9SWSFPTGGAVGYY186 pKa = 7.29FTSPAAANGEE196 pKa = 4.01IFVAGNEE203 pKa = 4.22SDD205 pKa = 4.42LSCIDD210 pKa = 3.3ITTHH214 pKa = 5.01TATWSVALPTAVKK227 pKa = 8.76STPAIGDD234 pKa = 3.33GKK236 pKa = 11.14VYY238 pKa = 9.58VTTAEE243 pKa = 3.82RR244 pKa = 11.84LYY246 pKa = 11.11ALSASTGAEE255 pKa = 3.7VWNASIGGTSSTPAVAGEE273 pKa = 4.1TVIAGSSDD281 pKa = 3.2GLHH284 pKa = 6.92AYY286 pKa = 9.6DD287 pKa = 5.18AGTGTPLWNFPSARR301 pKa = 11.84VDD303 pKa = 3.22VSPIIAGNLVYY314 pKa = 10.47AATNEE319 pKa = 4.34EE320 pKa = 4.25IGTVYY325 pKa = 10.74AVDD328 pKa = 3.26TGTGEE333 pKa = 4.42EE334 pKa = 4.29VWSYY338 pKa = 10.74TIEE341 pKa = 4.24APGDD345 pKa = 3.37GTFAAFFASSPAVSDD360 pKa = 3.53GVLYY364 pKa = 10.46IGVEE368 pKa = 4.05NNRR371 pKa = 11.84LYY373 pKa = 11.17AFGEE377 pKa = 4.74GSVSPTPTPTPTPTPAPGSWNGTVILAKK405 pKa = 9.54EE406 pKa = 4.44TFTFTPSNNASATYY420 pKa = 8.79TVNRR424 pKa = 11.84TTDD427 pKa = 3.49LGALTLAAQAGGFTINASDD446 pKa = 3.49AWYY449 pKa = 10.44ADD451 pKa = 3.35YY452 pKa = 11.19GSFMLEE458 pKa = 4.46DD459 pKa = 3.19IAGIANEE466 pKa = 4.9DD467 pKa = 3.25WTQEE471 pKa = 3.97NARR474 pKa = 11.84SWSIFINGAMAPAGLGANTLADD496 pKa = 3.82GDD498 pKa = 4.0RR499 pKa = 11.84LAFYY503 pKa = 9.66YY504 pKa = 10.65CPSDD508 pKa = 3.68PDD510 pKa = 3.68TYY512 pKa = 11.71APLIDD517 pKa = 3.44QAGYY521 pKa = 10.83VVTIDD526 pKa = 3.29VNVRR530 pKa = 11.84DD531 pKa = 4.58FTWEE535 pKa = 4.03GAVSLTDD542 pKa = 3.52GQTFTITPFNNEE554 pKa = 4.0SAIHH558 pKa = 5.12TFNRR562 pKa = 11.84TSALGALDD570 pKa = 3.82AAATAGGFNYY580 pKa = 9.48TVQEE584 pKa = 4.46TTWGPFLYY592 pKa = 10.46SIGGIAYY599 pKa = 10.29NEE601 pKa = 4.26TSWDD605 pKa = 3.08SWLYY609 pKa = 10.21SVNGIDD615 pKa = 4.45ASVGAADD622 pKa = 3.81YY623 pKa = 11.08QLTDD627 pKa = 3.82GDD629 pKa = 5.05VITYY633 pKa = 8.76WYY635 pKa = 9.08GAWGSTPDD643 pKa = 3.33TAGAVVDD650 pKa = 3.67ITVSIPATPAPTPTSGGGGGGGDD673 pKa = 3.87SPPSRR678 pKa = 11.84ITVTLQPGTFTITAEE693 pKa = 4.09NSGKK697 pKa = 9.38NHH699 pKa = 5.02TVSRR703 pKa = 11.84QTALGALDD711 pKa = 3.68ATGIAYY717 pKa = 8.52TIDD720 pKa = 3.16DD721 pKa = 4.42SYY723 pKa = 9.25YY724 pKa = 10.11QEE726 pKa = 4.39YY727 pKa = 11.0GSLFLSSIRR736 pKa = 11.84GRR738 pKa = 11.84VSEE741 pKa = 4.26GTRR744 pKa = 11.84GWMYY748 pKa = 10.47RR749 pKa = 11.84VNGGSPAVGANAYY762 pKa = 8.15PVTSGDD768 pKa = 3.48DD769 pKa = 3.82VIFFWSEE776 pKa = 4.09SMSSTPATSPDD787 pKa = 3.61VISIRR792 pKa = 11.84VVIPASSGSSGSDD805 pKa = 2.92SSGGGGSGSVPSSTTQEE822 pKa = 4.05QPNSVDD828 pKa = 3.09NSSASFLFGLPDD840 pKa = 3.61GAVIEE845 pKa = 4.3LGEE848 pKa = 4.05WGQTFSINTGPASAGEE864 pKa = 4.15EE865 pKa = 4.09VTISGNTFTINQSGIVLTIVARR887 pKa = 11.84NIDD890 pKa = 3.95EE891 pKa = 4.44KK892 pKa = 11.44DD893 pKa = 3.55GVASGLIEE901 pKa = 4.52SVTAAIDD908 pKa = 4.47LISGEE913 pKa = 4.03IEE915 pKa = 4.59GIGVVAASLDD925 pKa = 3.92LNLTGIPAADD935 pKa = 3.56GRR937 pKa = 11.84LDD939 pKa = 3.21ITFNATPDD947 pKa = 3.43ATAGNAFTLAATEE960 pKa = 3.88NDD962 pKa = 3.59EE963 pKa = 5.33EE964 pKa = 4.68IDD966 pKa = 3.5ALAYY970 pKa = 9.62TMTVTRR976 pKa = 11.84TNLEE980 pKa = 3.83NGEE983 pKa = 4.95DD984 pKa = 3.14ITGAVIRR991 pKa = 11.84MTINPEE997 pKa = 3.39WVEE1000 pKa = 3.72EE1001 pKa = 4.15HH1002 pKa = 6.59GGVDD1006 pKa = 3.24AVRR1009 pKa = 11.84IARR1012 pKa = 11.84SAEE1015 pKa = 3.92DD1016 pKa = 3.52STHH1019 pKa = 6.76EE1020 pKa = 4.36ILDD1023 pKa = 3.6TRR1025 pKa = 11.84LAGTDD1030 pKa = 3.39EE1031 pKa = 4.22NGNLIFEE1038 pKa = 4.54AVSPGGLSVFGLLTVRR1054 pKa = 11.84PASEE1058 pKa = 4.08MQEE1061 pKa = 4.41TPAVTATDD1069 pKa = 4.22PVSSTPVAAGTPEE1082 pKa = 4.19GAPPSLSSPFIGVGVGAALLIGAAYY1107 pKa = 10.23LIIRR1111 pKa = 11.84WRR1113 pKa = 11.84RR1114 pKa = 11.84EE1115 pKa = 3.43RR1116 pKa = 3.89
Molecular weight: 115.71 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0X3BJQ0|A0A0X3BJQ0_9EURY UPF0285 protein MMAB1_0915 OS=Methanoculleus bourgensis OX=83986 GN=MMAB1_0915 PE=3 SV=1
MM1 pKa = 7.16VSGVNSFLARR11 pKa = 11.84QSSGGCTRR19 pKa = 11.84VTKK22 pKa = 10.86NIMVLTAAQSNIILYY37 pKa = 7.95NTIAHH42 pKa = 5.93NEE44 pKa = 4.02VRR46 pKa = 11.84SSARR50 pKa = 11.84PLKK53 pKa = 9.85WFVNARR59 pKa = 11.84AAGTLALPQPGRR71 pKa = 11.84LLFQRR76 pKa = 11.84RR77 pKa = 11.84EE78 pKa = 4.09FVDD81 pKa = 3.76EE82 pKa = 4.14VVPSSRR88 pKa = 11.84YY89 pKa = 9.78ARR91 pKa = 11.84FLRR94 pKa = 11.84SSPPIDD100 pKa = 3.44KK101 pKa = 9.21TATPDD106 pKa = 3.37MPGDD110 pKa = 3.6QASDD114 pKa = 3.57PRR116 pKa = 11.84LGTAGSIARR125 pKa = 11.84CDD127 pKa = 3.8TIAVDD132 pKa = 4.14LRR134 pKa = 11.84RR135 pKa = 11.84HH136 pKa = 5.73EE137 pKa = 4.49GRR139 pKa = 11.84PCAGWKK145 pKa = 9.6
MM1 pKa = 7.16VSGVNSFLARR11 pKa = 11.84QSSGGCTRR19 pKa = 11.84VTKK22 pKa = 10.86NIMVLTAAQSNIILYY37 pKa = 7.95NTIAHH42 pKa = 5.93NEE44 pKa = 4.02VRR46 pKa = 11.84SSARR50 pKa = 11.84PLKK53 pKa = 9.85WFVNARR59 pKa = 11.84AAGTLALPQPGRR71 pKa = 11.84LLFQRR76 pKa = 11.84RR77 pKa = 11.84EE78 pKa = 4.09FVDD81 pKa = 3.76EE82 pKa = 4.14VVPSSRR88 pKa = 11.84YY89 pKa = 9.78ARR91 pKa = 11.84FLRR94 pKa = 11.84SSPPIDD100 pKa = 3.44KK101 pKa = 9.21TATPDD106 pKa = 3.37MPGDD110 pKa = 3.6QASDD114 pKa = 3.57PRR116 pKa = 11.84LGTAGSIARR125 pKa = 11.84CDD127 pKa = 3.8TIAVDD132 pKa = 4.14LRR134 pKa = 11.84RR135 pKa = 11.84HH136 pKa = 5.73EE137 pKa = 4.49GRR139 pKa = 11.84PCAGWKK145 pKa = 9.6
Molecular weight: 15.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
822921 |
13 |
2024 |
238.8 |
26.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.62 ± 0.059 | 1.352 ± 0.022 |
5.658 ± 0.04 | 6.787 ± 0.049 |
3.538 ± 0.028 | 8.471 ± 0.044 |
2.025 ± 0.022 | 6.218 ± 0.037 |
3.402 ± 0.039 | 9.219 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.446 ± 0.025 | 2.767 ± 0.033 |
5.259 ± 0.034 | 2.537 ± 0.024 |
7.364 ± 0.066 | 5.569 ± 0.046 |
5.702 ± 0.058 | 7.896 ± 0.039 |
1.141 ± 0.018 | 3.028 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
