
Diporeia-associated CRESS-DNA virus LH481
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 8.84
Get precalculated fractions of proteins
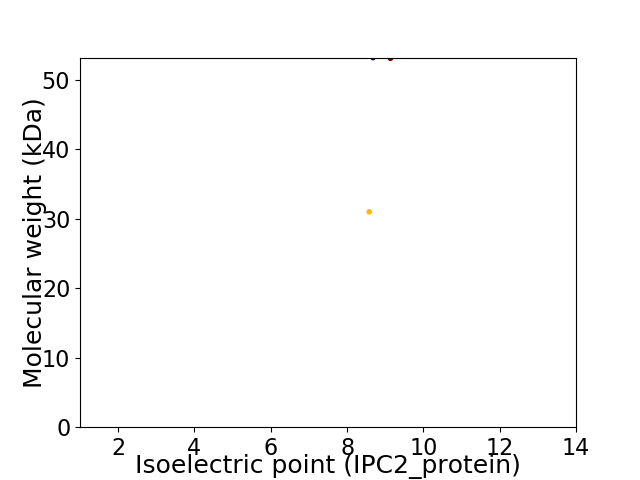
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
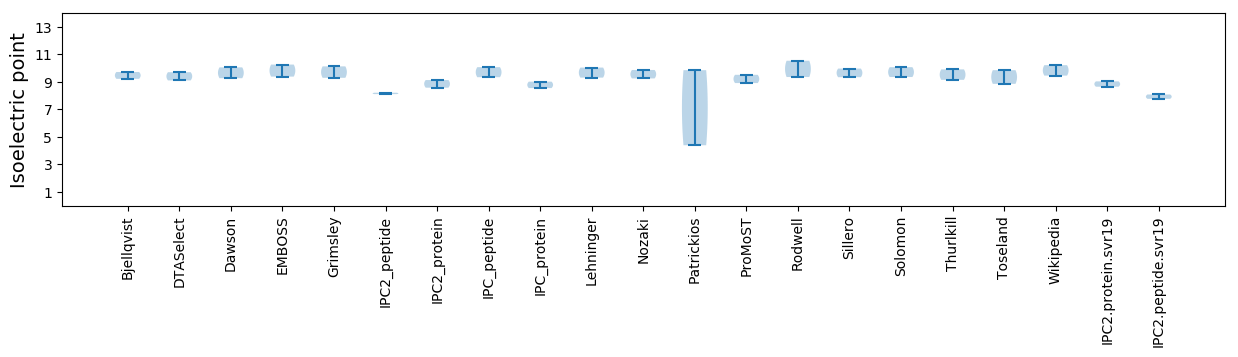
Protein with the lowest isoelectric point:
>tr|A0A1X8VHH5|A0A1X8VHH5_9VIRU Putative capsid protein OS=Diporeia-associated CRESS-DNA virus LH481 OX=1974422 GN=Cap PE=4 SV=1
MM1 pKa = 7.25TIKK4 pKa = 9.35MQNGQGTYY12 pKa = 9.55WILTIPRR19 pKa = 11.84EE20 pKa = 4.18SFVPYY25 pKa = 10.07LPPGIAYY32 pKa = 9.53IKK34 pKa = 10.14GQMEE38 pKa = 4.44IGASGYY44 pKa = 7.94NHH46 pKa = 6.13WQIIAICDD54 pKa = 3.27RR55 pKa = 11.84SRR57 pKa = 11.84RR58 pKa = 11.84LSWIRR63 pKa = 11.84SIFGNFHH70 pKa = 6.87AEE72 pKa = 4.01LTRR75 pKa = 11.84SVAAEE80 pKa = 3.73QYY82 pKa = 8.54VWKK85 pKa = 10.65DD86 pKa = 3.25EE87 pKa = 4.22TAVVGTRR94 pKa = 11.84FEE96 pKa = 5.02LGSKK100 pKa = 9.7PIRR103 pKa = 11.84RR104 pKa = 11.84NAKK107 pKa = 9.81KK108 pKa = 10.24DD109 pKa = 2.77WDD111 pKa = 5.01AIWSSACSGDD121 pKa = 3.31ILAIPSDD128 pKa = 3.65VRR130 pKa = 11.84IRR132 pKa = 11.84CYY134 pKa = 10.0STLKK138 pKa = 10.49RR139 pKa = 11.84IRR141 pKa = 11.84ADD143 pKa = 3.29YY144 pKa = 10.56LEE146 pKa = 4.63PVSMVRR152 pKa = 11.84SCTVFWGPTGTGKK165 pKa = 10.79SMRR168 pKa = 11.84SWNEE172 pKa = 3.28AGLQAYY178 pKa = 8.1PKK180 pKa = 10.65DD181 pKa = 3.71PNTKK185 pKa = 9.1FWCGYY190 pKa = 9.04SGQEE194 pKa = 3.62HH195 pKa = 5.24VVIDD199 pKa = 3.85EE200 pKa = 4.08FRR202 pKa = 11.84GKK204 pKa = 9.97IDD206 pKa = 3.24ISHH209 pKa = 7.64LLRR212 pKa = 11.84WLDD215 pKa = 3.65RR216 pKa = 11.84YY217 pKa = 9.3PVRR220 pKa = 11.84VEE222 pKa = 3.91LKK224 pKa = 9.7GASFPLMATKK234 pKa = 10.53YY235 pKa = 8.8WITSNLHH242 pKa = 4.74PRR244 pKa = 11.84EE245 pKa = 4.3WYY247 pKa = 9.92PEE249 pKa = 3.68IDD251 pKa = 3.5AYY253 pKa = 10.05TYY255 pKa = 10.65DD256 pKa = 3.43ALEE259 pKa = 4.06RR260 pKa = 11.84RR261 pKa = 11.84LNIINITT268 pKa = 3.55
MM1 pKa = 7.25TIKK4 pKa = 9.35MQNGQGTYY12 pKa = 9.55WILTIPRR19 pKa = 11.84EE20 pKa = 4.18SFVPYY25 pKa = 10.07LPPGIAYY32 pKa = 9.53IKK34 pKa = 10.14GQMEE38 pKa = 4.44IGASGYY44 pKa = 7.94NHH46 pKa = 6.13WQIIAICDD54 pKa = 3.27RR55 pKa = 11.84SRR57 pKa = 11.84RR58 pKa = 11.84LSWIRR63 pKa = 11.84SIFGNFHH70 pKa = 6.87AEE72 pKa = 4.01LTRR75 pKa = 11.84SVAAEE80 pKa = 3.73QYY82 pKa = 8.54VWKK85 pKa = 10.65DD86 pKa = 3.25EE87 pKa = 4.22TAVVGTRR94 pKa = 11.84FEE96 pKa = 5.02LGSKK100 pKa = 9.7PIRR103 pKa = 11.84RR104 pKa = 11.84NAKK107 pKa = 9.81KK108 pKa = 10.24DD109 pKa = 2.77WDD111 pKa = 5.01AIWSSACSGDD121 pKa = 3.31ILAIPSDD128 pKa = 3.65VRR130 pKa = 11.84IRR132 pKa = 11.84CYY134 pKa = 10.0STLKK138 pKa = 10.49RR139 pKa = 11.84IRR141 pKa = 11.84ADD143 pKa = 3.29YY144 pKa = 10.56LEE146 pKa = 4.63PVSMVRR152 pKa = 11.84SCTVFWGPTGTGKK165 pKa = 10.79SMRR168 pKa = 11.84SWNEE172 pKa = 3.28AGLQAYY178 pKa = 8.1PKK180 pKa = 10.65DD181 pKa = 3.71PNTKK185 pKa = 9.1FWCGYY190 pKa = 9.04SGQEE194 pKa = 3.62HH195 pKa = 5.24VVIDD199 pKa = 3.85EE200 pKa = 4.08FRR202 pKa = 11.84GKK204 pKa = 9.97IDD206 pKa = 3.24ISHH209 pKa = 7.64LLRR212 pKa = 11.84WLDD215 pKa = 3.65RR216 pKa = 11.84YY217 pKa = 9.3PVRR220 pKa = 11.84VEE222 pKa = 3.91LKK224 pKa = 9.7GASFPLMATKK234 pKa = 10.53YY235 pKa = 8.8WITSNLHH242 pKa = 4.74PRR244 pKa = 11.84EE245 pKa = 4.3WYY247 pKa = 9.92PEE249 pKa = 3.68IDD251 pKa = 3.5AYY253 pKa = 10.05TYY255 pKa = 10.65DD256 pKa = 3.43ALEE259 pKa = 4.06RR260 pKa = 11.84RR261 pKa = 11.84LNIINITT268 pKa = 3.55
Molecular weight: 31.0 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X8VHH5|A0A1X8VHH5_9VIRU Putative capsid protein OS=Diporeia-associated CRESS-DNA virus LH481 OX=1974422 GN=Cap PE=4 SV=1
MM1 pKa = 7.26GLKK4 pKa = 10.25RR5 pKa = 11.84KK6 pKa = 9.0IANAAGATLGFIAGDD21 pKa = 3.3IPGAMTGADD30 pKa = 4.14LADD33 pKa = 4.35SLYY36 pKa = 10.18KK37 pKa = 9.72WKK39 pKa = 11.14YY40 pKa = 8.35PDD42 pKa = 3.25QKK44 pKa = 11.47NFAEE48 pKa = 4.9KK49 pKa = 10.39INMGKK54 pKa = 9.4YY55 pKa = 9.37VKK57 pKa = 10.38SGSKK61 pKa = 10.1RR62 pKa = 11.84KK63 pKa = 9.61ASKK66 pKa = 10.12QSGGKK71 pKa = 8.5RR72 pKa = 11.84RR73 pKa = 11.84KK74 pKa = 8.12VSKK77 pKa = 10.71YY78 pKa = 9.57SGNKK82 pKa = 9.24KK83 pKa = 8.89VEE85 pKa = 3.85KK86 pKa = 9.79MRR88 pKa = 11.84NNNMNQTNLTNTHH101 pKa = 6.0IKK103 pKa = 10.63KK104 pKa = 10.22KK105 pKa = 9.68DD106 pKa = 3.39TLKK109 pKa = 9.99MRR111 pKa = 11.84KK112 pKa = 8.73SPYY115 pKa = 9.13KK116 pKa = 10.39VRR118 pKa = 11.84VSSAFRR124 pKa = 11.84KK125 pKa = 9.72KK126 pKa = 10.36VIKK129 pKa = 10.28AVSDD133 pKa = 3.77KK134 pKa = 10.58EE135 pKa = 4.35PQGSATLTFIGGFFPPNGNVGWPVRR160 pKa = 11.84DD161 pKa = 3.58SVSKK165 pKa = 10.19QWVFGLPMGNQATSATAYY183 pKa = 10.99SNGANIRR190 pKa = 11.84SGDD193 pKa = 3.73VFRR196 pKa = 11.84TNMLLHH202 pKa = 5.93VASRR206 pKa = 11.84MWNGAAKK213 pKa = 8.83ATNFNEE219 pKa = 4.01FGYY222 pKa = 10.97DD223 pKa = 3.77RR224 pKa = 11.84LDD226 pKa = 3.52NFCNPEE232 pKa = 4.0TSVGALKK239 pKa = 10.76LDD241 pKa = 3.75ILKK244 pKa = 9.94SWAVIEE250 pKa = 4.33MKK252 pKa = 10.72NVTRR256 pKa = 11.84TKK258 pKa = 10.47LRR260 pKa = 11.84LYY262 pKa = 10.18MYY264 pKa = 9.71VCKK267 pKa = 10.1PKK269 pKa = 9.63YY270 pKa = 8.7QRR272 pKa = 11.84RR273 pKa = 11.84TADD276 pKa = 3.03FNARR280 pKa = 11.84VDD282 pKa = 4.0GVNSGIPTQFTEE294 pKa = 4.33FGTNISGVQASTYY307 pKa = 10.76GIFPEE312 pKa = 4.54MTPAWKK318 pKa = 9.92RR319 pKa = 11.84QWEE322 pKa = 4.24FEE324 pKa = 3.94KK325 pKa = 11.38YY326 pKa = 10.3EE327 pKa = 4.02IVLEE331 pKa = 4.2PGQEE335 pKa = 3.84HH336 pKa = 6.45RR337 pKa = 11.84HH338 pKa = 5.66TINGYY343 pKa = 9.52CGQFDD348 pKa = 3.82FLKK351 pKa = 10.24AYY353 pKa = 9.59KK354 pKa = 10.41DD355 pKa = 3.68SVHH358 pKa = 6.73QNVQPIDD365 pKa = 3.34RR366 pKa = 11.84HH367 pKa = 5.04VFFVMQTDD375 pKa = 3.89LGIVRR380 pKa = 11.84EE381 pKa = 4.33GTPAVSYY388 pKa = 9.66GVRR391 pKa = 11.84TAQDD395 pKa = 3.1SDD397 pKa = 3.97DD398 pKa = 4.03ASLGLGILFEE408 pKa = 4.75TKK410 pKa = 9.5VHH412 pKa = 5.86WTLQMPEE419 pKa = 3.86PAGARR424 pKa = 11.84MPATGLLGVGAYY436 pKa = 8.19NKK438 pKa = 9.75PAGFRR443 pKa = 11.84KK444 pKa = 9.57RR445 pKa = 11.84CYY447 pKa = 10.31LYY449 pKa = 10.49DD450 pKa = 5.27IIDD453 pKa = 4.34DD454 pKa = 3.92TDD456 pKa = 4.02LPNQTEE462 pKa = 4.47VVNIDD467 pKa = 3.74DD468 pKa = 4.27NNPTVVVAA476 pKa = 5.12
MM1 pKa = 7.26GLKK4 pKa = 10.25RR5 pKa = 11.84KK6 pKa = 9.0IANAAGATLGFIAGDD21 pKa = 3.3IPGAMTGADD30 pKa = 4.14LADD33 pKa = 4.35SLYY36 pKa = 10.18KK37 pKa = 9.72WKK39 pKa = 11.14YY40 pKa = 8.35PDD42 pKa = 3.25QKK44 pKa = 11.47NFAEE48 pKa = 4.9KK49 pKa = 10.39INMGKK54 pKa = 9.4YY55 pKa = 9.37VKK57 pKa = 10.38SGSKK61 pKa = 10.1RR62 pKa = 11.84KK63 pKa = 9.61ASKK66 pKa = 10.12QSGGKK71 pKa = 8.5RR72 pKa = 11.84RR73 pKa = 11.84KK74 pKa = 8.12VSKK77 pKa = 10.71YY78 pKa = 9.57SGNKK82 pKa = 9.24KK83 pKa = 8.89VEE85 pKa = 3.85KK86 pKa = 9.79MRR88 pKa = 11.84NNNMNQTNLTNTHH101 pKa = 6.0IKK103 pKa = 10.63KK104 pKa = 10.22KK105 pKa = 9.68DD106 pKa = 3.39TLKK109 pKa = 9.99MRR111 pKa = 11.84KK112 pKa = 8.73SPYY115 pKa = 9.13KK116 pKa = 10.39VRR118 pKa = 11.84VSSAFRR124 pKa = 11.84KK125 pKa = 9.72KK126 pKa = 10.36VIKK129 pKa = 10.28AVSDD133 pKa = 3.77KK134 pKa = 10.58EE135 pKa = 4.35PQGSATLTFIGGFFPPNGNVGWPVRR160 pKa = 11.84DD161 pKa = 3.58SVSKK165 pKa = 10.19QWVFGLPMGNQATSATAYY183 pKa = 10.99SNGANIRR190 pKa = 11.84SGDD193 pKa = 3.73VFRR196 pKa = 11.84TNMLLHH202 pKa = 5.93VASRR206 pKa = 11.84MWNGAAKK213 pKa = 8.83ATNFNEE219 pKa = 4.01FGYY222 pKa = 10.97DD223 pKa = 3.77RR224 pKa = 11.84LDD226 pKa = 3.52NFCNPEE232 pKa = 4.0TSVGALKK239 pKa = 10.76LDD241 pKa = 3.75ILKK244 pKa = 9.94SWAVIEE250 pKa = 4.33MKK252 pKa = 10.72NVTRR256 pKa = 11.84TKK258 pKa = 10.47LRR260 pKa = 11.84LYY262 pKa = 10.18MYY264 pKa = 9.71VCKK267 pKa = 10.1PKK269 pKa = 9.63YY270 pKa = 8.7QRR272 pKa = 11.84RR273 pKa = 11.84TADD276 pKa = 3.03FNARR280 pKa = 11.84VDD282 pKa = 4.0GVNSGIPTQFTEE294 pKa = 4.33FGTNISGVQASTYY307 pKa = 10.76GIFPEE312 pKa = 4.54MTPAWKK318 pKa = 9.92RR319 pKa = 11.84QWEE322 pKa = 4.24FEE324 pKa = 3.94KK325 pKa = 11.38YY326 pKa = 10.3EE327 pKa = 4.02IVLEE331 pKa = 4.2PGQEE335 pKa = 3.84HH336 pKa = 6.45RR337 pKa = 11.84HH338 pKa = 5.66TINGYY343 pKa = 9.52CGQFDD348 pKa = 3.82FLKK351 pKa = 10.24AYY353 pKa = 9.59KK354 pKa = 10.41DD355 pKa = 3.68SVHH358 pKa = 6.73QNVQPIDD365 pKa = 3.34RR366 pKa = 11.84HH367 pKa = 5.04VFFVMQTDD375 pKa = 3.89LGIVRR380 pKa = 11.84EE381 pKa = 4.33GTPAVSYY388 pKa = 9.66GVRR391 pKa = 11.84TAQDD395 pKa = 3.1SDD397 pKa = 3.97DD398 pKa = 4.03ASLGLGILFEE408 pKa = 4.75TKK410 pKa = 9.5VHH412 pKa = 5.86WTLQMPEE419 pKa = 3.86PAGARR424 pKa = 11.84MPATGLLGVGAYY436 pKa = 8.19NKK438 pKa = 9.75PAGFRR443 pKa = 11.84KK444 pKa = 9.57RR445 pKa = 11.84CYY447 pKa = 10.31LYY449 pKa = 10.49DD450 pKa = 5.27IIDD453 pKa = 4.34DD454 pKa = 3.92TDD456 pKa = 4.02LPNQTEE462 pKa = 4.47VVNIDD467 pKa = 3.74DD468 pKa = 4.27NNPTVVVAA476 pKa = 5.12
Molecular weight: 53.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
744 |
268 |
476 |
372.0 |
42.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.392 ± 0.404 | 1.21 ± 0.392 |
5.242 ± 0.234 | 4.167 ± 0.631 |
4.032 ± 0.625 | 7.93 ± 0.725 |
1.613 ± 0.151 | 6.183 ± 1.878 |
7.392 ± 1.518 | 5.914 ± 0.256 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.823 ± 0.349 | 5.511 ± 1.285 |
4.839 ± 0.23 | 3.36 ± 0.447 |
6.452 ± 1.049 | 6.317 ± 0.684 |
6.183 ± 0.35 | 6.452 ± 0.956 |
2.688 ± 1.069 | 4.301 ± 0.551 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
