
Paracoccus phage vB_PsuS_Psul1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 7.11
Get precalculated fractions of proteins
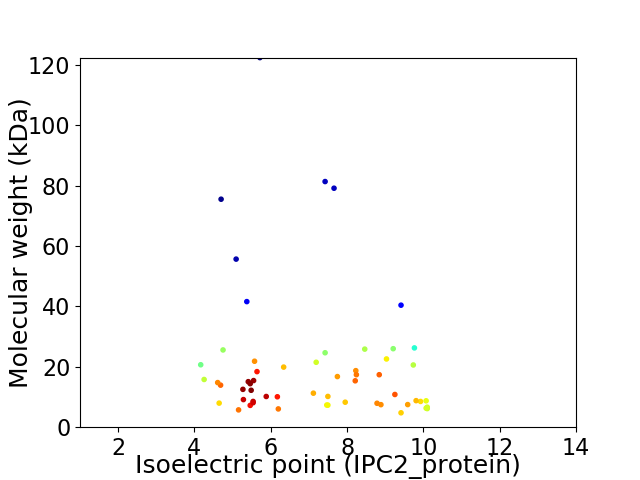
Virtual 2D-PAGE plot for 57 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3T0IK20|A0A3T0IK20_9CAUD Scaffolding protein OS=Paracoccus phage vB_PsuS_Psul1 OX=2500569 GN=psul1_p31 PE=4 SV=1
MM1 pKa = 7.43SDD3 pKa = 2.88KK4 pKa = 10.28VTVYY8 pKa = 10.04EE9 pKa = 4.18AATGKK14 pKa = 9.53IRR16 pKa = 11.84RR17 pKa = 11.84VLLLPLVGMLDD28 pKa = 3.63EE29 pKa = 5.55DD30 pKa = 4.65GEE32 pKa = 4.73IVGHH36 pKa = 7.09IINPVDD42 pKa = 3.82LANNVAGDD50 pKa = 3.56EE51 pKa = 4.26RR52 pKa = 11.84YY53 pKa = 10.41VLGEE57 pKa = 3.89FGSDD61 pKa = 2.48RR62 pKa = 11.84FYY64 pKa = 11.05WDD66 pKa = 3.61GSAMAAFPDD75 pKa = 4.79PPSEE79 pKa = 3.67WAEE82 pKa = 3.6FDD84 pKa = 3.34FAARR88 pKa = 11.84QWVDD92 pKa = 2.83HH93 pKa = 6.5YY94 pKa = 11.89DD95 pKa = 3.59PALALAAARR104 pKa = 11.84ASANMTRR111 pKa = 11.84SDD113 pKa = 3.92FLLAAISAGIILPSDD128 pKa = 4.09AGPAARR134 pKa = 11.84GEE136 pKa = 4.31IPPSLVPVFDD146 pKa = 4.15GLPLEE151 pKa = 4.32VRR153 pKa = 11.84IEE155 pKa = 3.97AVVRR159 pKa = 11.84WGAATVIEE167 pKa = 4.34RR168 pKa = 11.84TNPVLAALAEE178 pKa = 4.2AMAITDD184 pKa = 4.08ADD186 pKa = 4.04LDD188 pKa = 4.01ALFGISS194 pKa = 3.49
MM1 pKa = 7.43SDD3 pKa = 2.88KK4 pKa = 10.28VTVYY8 pKa = 10.04EE9 pKa = 4.18AATGKK14 pKa = 9.53IRR16 pKa = 11.84RR17 pKa = 11.84VLLLPLVGMLDD28 pKa = 3.63EE29 pKa = 5.55DD30 pKa = 4.65GEE32 pKa = 4.73IVGHH36 pKa = 7.09IINPVDD42 pKa = 3.82LANNVAGDD50 pKa = 3.56EE51 pKa = 4.26RR52 pKa = 11.84YY53 pKa = 10.41VLGEE57 pKa = 3.89FGSDD61 pKa = 2.48RR62 pKa = 11.84FYY64 pKa = 11.05WDD66 pKa = 3.61GSAMAAFPDD75 pKa = 4.79PPSEE79 pKa = 3.67WAEE82 pKa = 3.6FDD84 pKa = 3.34FAARR88 pKa = 11.84QWVDD92 pKa = 2.83HH93 pKa = 6.5YY94 pKa = 11.89DD95 pKa = 3.59PALALAAARR104 pKa = 11.84ASANMTRR111 pKa = 11.84SDD113 pKa = 3.92FLLAAISAGIILPSDD128 pKa = 4.09AGPAARR134 pKa = 11.84GEE136 pKa = 4.31IPPSLVPVFDD146 pKa = 4.15GLPLEE151 pKa = 4.32VRR153 pKa = 11.84IEE155 pKa = 3.97AVVRR159 pKa = 11.84WGAATVIEE167 pKa = 4.34RR168 pKa = 11.84TNPVLAALAEE178 pKa = 4.2AMAITDD184 pKa = 4.08ADD186 pKa = 4.04LDD188 pKa = 4.01ALFGISS194 pKa = 3.49
Molecular weight: 20.68 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3Q9R8W0|A0A3Q9R8W0_9CAUD Putative replication protein OS=Paracoccus phage vB_PsuS_Psul1 OX=2500569 GN=psul1_p21 PE=4 SV=1
MM1 pKa = 7.18TKK3 pKa = 10.6AFRR6 pKa = 11.84EE7 pKa = 3.96AAAARR12 pKa = 11.84IAAEE16 pKa = 4.24RR17 pKa = 11.84ARR19 pKa = 11.84LDD21 pKa = 4.52AILHH25 pKa = 6.2RR26 pKa = 11.84ATPVDD31 pKa = 3.33GCGPAIPIAPARR43 pKa = 11.84GPQIVVTPRR52 pKa = 11.84RR53 pKa = 11.84MVPDD57 pKa = 3.96AKK59 pKa = 10.75DD60 pKa = 3.17ATGWKK65 pKa = 9.88VEE67 pKa = 3.97EE68 pKa = 4.37LGWRR72 pKa = 11.84GFNAVRR78 pKa = 11.84ATDD81 pKa = 3.36IFDD84 pKa = 4.12DD85 pKa = 4.43LARR88 pKa = 11.84RR89 pKa = 11.84AAKK92 pKa = 10.34RR93 pKa = 11.84KK94 pKa = 9.02DD95 pKa = 3.56APPFTPGQVAVGRR108 pKa = 11.84LYY110 pKa = 10.27RR111 pKa = 11.84DD112 pKa = 2.95LVEE115 pKa = 4.22RR116 pKa = 11.84HH117 pKa = 6.43DD118 pKa = 4.26AGGMRR123 pKa = 11.84CASLEE128 pKa = 3.94AGRR131 pKa = 11.84GGSPSGGGEE140 pKa = 4.05FIDD143 pKa = 4.5AFIEE147 pKa = 3.93EE148 pKa = 4.36GRR150 pKa = 11.84VIEE153 pKa = 4.01LLRR156 pKa = 11.84RR157 pKa = 11.84RR158 pKa = 11.84IGHH161 pKa = 5.9GVAMPVRR168 pKa = 11.84RR169 pKa = 11.84VRR171 pKa = 11.84PSARR175 pKa = 11.84GGDD178 pKa = 4.27GARR181 pKa = 11.84MIHH184 pKa = 6.93DD185 pKa = 3.89RR186 pKa = 11.84VLVDD190 pKa = 5.73DD191 pKa = 4.2ICLHH195 pKa = 5.65GRR197 pKa = 11.84SFQSVLEE204 pKa = 3.95RR205 pKa = 11.84HH206 pKa = 5.85GWTKK210 pKa = 9.95TGRR213 pKa = 11.84NVVALQCALAACLDD227 pKa = 3.53RR228 pKa = 11.84MQFHH232 pKa = 7.13SKK234 pKa = 9.5MGLTTT239 pKa = 3.72
MM1 pKa = 7.18TKK3 pKa = 10.6AFRR6 pKa = 11.84EE7 pKa = 3.96AAAARR12 pKa = 11.84IAAEE16 pKa = 4.24RR17 pKa = 11.84ARR19 pKa = 11.84LDD21 pKa = 4.52AILHH25 pKa = 6.2RR26 pKa = 11.84ATPVDD31 pKa = 3.33GCGPAIPIAPARR43 pKa = 11.84GPQIVVTPRR52 pKa = 11.84RR53 pKa = 11.84MVPDD57 pKa = 3.96AKK59 pKa = 10.75DD60 pKa = 3.17ATGWKK65 pKa = 9.88VEE67 pKa = 3.97EE68 pKa = 4.37LGWRR72 pKa = 11.84GFNAVRR78 pKa = 11.84ATDD81 pKa = 3.36IFDD84 pKa = 4.12DD85 pKa = 4.43LARR88 pKa = 11.84RR89 pKa = 11.84AAKK92 pKa = 10.34RR93 pKa = 11.84KK94 pKa = 9.02DD95 pKa = 3.56APPFTPGQVAVGRR108 pKa = 11.84LYY110 pKa = 10.27RR111 pKa = 11.84DD112 pKa = 2.95LVEE115 pKa = 4.22RR116 pKa = 11.84HH117 pKa = 6.43DD118 pKa = 4.26AGGMRR123 pKa = 11.84CASLEE128 pKa = 3.94AGRR131 pKa = 11.84GGSPSGGGEE140 pKa = 4.05FIDD143 pKa = 4.5AFIEE147 pKa = 3.93EE148 pKa = 4.36GRR150 pKa = 11.84VIEE153 pKa = 4.01LLRR156 pKa = 11.84RR157 pKa = 11.84RR158 pKa = 11.84IGHH161 pKa = 5.9GVAMPVRR168 pKa = 11.84RR169 pKa = 11.84VRR171 pKa = 11.84PSARR175 pKa = 11.84GGDD178 pKa = 4.27GARR181 pKa = 11.84MIHH184 pKa = 6.93DD185 pKa = 3.89RR186 pKa = 11.84VLVDD190 pKa = 5.73DD191 pKa = 4.2ICLHH195 pKa = 5.65GRR197 pKa = 11.84SFQSVLEE204 pKa = 3.95RR205 pKa = 11.84HH206 pKa = 5.85GWTKK210 pKa = 9.95TGRR213 pKa = 11.84NVVALQCALAACLDD227 pKa = 3.53RR228 pKa = 11.84MQFHH232 pKa = 7.13SKK234 pKa = 9.5MGLTTT239 pKa = 3.72
Molecular weight: 26.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
10707 |
41 |
1143 |
187.8 |
20.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.795 ± 0.498 | 0.897 ± 0.158 |
5.875 ± 0.241 | 6.024 ± 0.427 |
3.269 ± 0.17 | 8.396 ± 0.33 |
2.008 ± 0.216 | 5.053 ± 0.226 |
3.699 ± 0.255 | 8.499 ± 0.307 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.933 ± 0.157 | 2.568 ± 0.184 |
5.286 ± 0.338 | 3.456 ± 0.18 |
8.154 ± 0.495 | 5.912 ± 0.24 |
5.333 ± 0.317 | 6.005 ± 0.317 |
1.933 ± 0.149 | 1.905 ± 0.186 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
