
Vibrio phage phiV039C
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; unclassified Myoviridae
Average proteome isoelectric point is 5.88
Get precalculated fractions of proteins
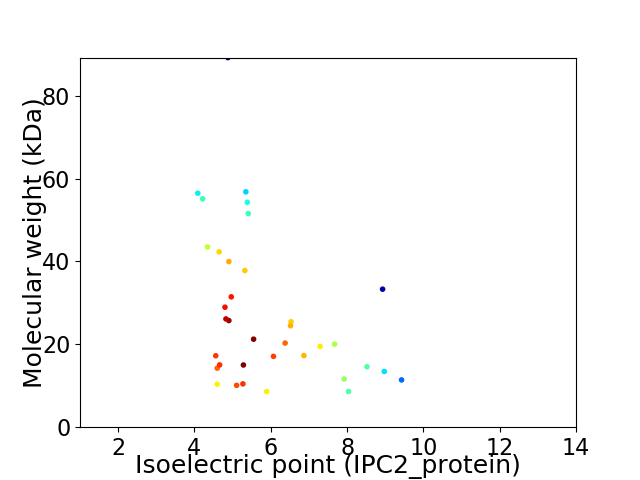
Virtual 2D-PAGE plot for 36 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6M3T8F1|A0A6M3T8F1_9CAUD Uncharacterized protein OS=Vibrio phage phiV039C OX=2705110 PE=4 SV=1
MM1 pKa = 7.0STIITADD8 pKa = 3.15GGNISSDD15 pKa = 3.23RR16 pKa = 11.84YY17 pKa = 9.82VEE19 pKa = 4.41IISGVGGAAAFPTRR33 pKa = 11.84QLILRR38 pKa = 11.84LFTNNEE44 pKa = 3.87AFPTNSVVTFSSPEE58 pKa = 3.81DD59 pKa = 3.34LLTYY63 pKa = 10.95LNGDD67 pKa = 3.75STAEE71 pKa = 4.03EE72 pKa = 4.47YY73 pKa = 10.5KK74 pKa = 10.06QAVYY78 pKa = 10.45YY79 pKa = 9.9FSYY82 pKa = 9.92ISKK85 pKa = 10.62AIRR88 pKa = 11.84TPKK91 pKa = 9.74QLSIARR97 pKa = 11.84WAEE100 pKa = 4.11VNTSAQVFGSKK111 pKa = 10.19AATLDD116 pKa = 3.3QLKK119 pKa = 9.37TYY121 pKa = 7.58TTATLDD127 pKa = 3.3VTLNGTTYY135 pKa = 10.93NATAIDD141 pKa = 4.37LSLAASFADD150 pKa = 3.97VATTLQGKK158 pKa = 9.6ISALDD163 pKa = 3.56ASLTAATVTYY173 pKa = 10.29NSASGGFDD181 pKa = 2.98VDD183 pKa = 4.06TNGDD187 pKa = 3.42ADD189 pKa = 3.91GALTFASATAGFLSDD204 pKa = 4.16IGLDD208 pKa = 3.45STAVFSSGIAEE219 pKa = 4.08QTLTEE224 pKa = 4.56LMTSSTNLSNNFGSLAFVQDD244 pKa = 4.06LSLTEE249 pKa = 4.02IEE251 pKa = 6.22EE252 pKa = 4.14VATWNKK258 pKa = 8.91GQNFAYY264 pKa = 8.94MYY266 pKa = 10.08CEE268 pKa = 4.26KK269 pKa = 9.97STKK272 pKa = 10.42ANSQSYY278 pKa = 9.61FDD280 pKa = 3.78TLKK283 pKa = 10.94GYY285 pKa = 10.83GGLAVTLYY293 pKa = 10.44DD294 pKa = 4.66DD295 pKa = 4.45SVADD299 pKa = 4.48EE300 pKa = 4.71YY301 pKa = 11.1PWLHH305 pKa = 6.63PAAEE309 pKa = 4.2TAAIDD314 pKa = 3.85PSKK317 pKa = 11.08PNAFPNYY324 pKa = 8.79MFAPDD329 pKa = 3.66SALSAVVTTDD339 pKa = 4.09SEE341 pKa = 4.87ADD343 pKa = 3.27TYY345 pKa = 10.98DD346 pKa = 3.58ARR348 pKa = 11.84RR349 pKa = 11.84MNYY352 pKa = 8.76MGRR355 pKa = 11.84TQEE358 pKa = 4.06AGQTRR363 pKa = 11.84TWYY366 pKa = 9.93QRR368 pKa = 11.84GVLMGGDD375 pKa = 3.57TDD377 pKa = 3.69ATTMSVYY384 pKa = 10.04MGEE387 pKa = 4.07AWLKK391 pKa = 11.26AEE393 pKa = 5.35LKK395 pKa = 10.64SQFLNMFNALPGITPDD411 pKa = 2.95IAGRR415 pKa = 11.84SYY417 pKa = 11.31INLYY421 pKa = 10.38LDD423 pKa = 3.67AAVAQALPDD432 pKa = 4.06GGNGMISVGKK442 pKa = 10.0QLTTTQQAYY451 pKa = 6.65ITQVSGDD458 pKa = 3.43EE459 pKa = 4.35DD460 pKa = 2.96AWKK463 pKa = 9.99QVQSEE468 pKa = 5.23GYY470 pKa = 8.11WYY472 pKa = 9.21TVNFFSTIAEE482 pKa = 4.18SGVTEE487 pKa = 4.22WTAEE491 pKa = 3.84YY492 pKa = 10.1TLIYY496 pKa = 9.42ATAEE500 pKa = 4.17KK501 pKa = 10.05VRR503 pKa = 11.84KK504 pKa = 9.94VSGTHH509 pKa = 5.91ILII512 pKa = 4.45
MM1 pKa = 7.0STIITADD8 pKa = 3.15GGNISSDD15 pKa = 3.23RR16 pKa = 11.84YY17 pKa = 9.82VEE19 pKa = 4.41IISGVGGAAAFPTRR33 pKa = 11.84QLILRR38 pKa = 11.84LFTNNEE44 pKa = 3.87AFPTNSVVTFSSPEE58 pKa = 3.81DD59 pKa = 3.34LLTYY63 pKa = 10.95LNGDD67 pKa = 3.75STAEE71 pKa = 4.03EE72 pKa = 4.47YY73 pKa = 10.5KK74 pKa = 10.06QAVYY78 pKa = 10.45YY79 pKa = 9.9FSYY82 pKa = 9.92ISKK85 pKa = 10.62AIRR88 pKa = 11.84TPKK91 pKa = 9.74QLSIARR97 pKa = 11.84WAEE100 pKa = 4.11VNTSAQVFGSKK111 pKa = 10.19AATLDD116 pKa = 3.3QLKK119 pKa = 9.37TYY121 pKa = 7.58TTATLDD127 pKa = 3.3VTLNGTTYY135 pKa = 10.93NATAIDD141 pKa = 4.37LSLAASFADD150 pKa = 3.97VATTLQGKK158 pKa = 9.6ISALDD163 pKa = 3.56ASLTAATVTYY173 pKa = 10.29NSASGGFDD181 pKa = 2.98VDD183 pKa = 4.06TNGDD187 pKa = 3.42ADD189 pKa = 3.91GALTFASATAGFLSDD204 pKa = 4.16IGLDD208 pKa = 3.45STAVFSSGIAEE219 pKa = 4.08QTLTEE224 pKa = 4.56LMTSSTNLSNNFGSLAFVQDD244 pKa = 4.06LSLTEE249 pKa = 4.02IEE251 pKa = 6.22EE252 pKa = 4.14VATWNKK258 pKa = 8.91GQNFAYY264 pKa = 8.94MYY266 pKa = 10.08CEE268 pKa = 4.26KK269 pKa = 9.97STKK272 pKa = 10.42ANSQSYY278 pKa = 9.61FDD280 pKa = 3.78TLKK283 pKa = 10.94GYY285 pKa = 10.83GGLAVTLYY293 pKa = 10.44DD294 pKa = 4.66DD295 pKa = 4.45SVADD299 pKa = 4.48EE300 pKa = 4.71YY301 pKa = 11.1PWLHH305 pKa = 6.63PAAEE309 pKa = 4.2TAAIDD314 pKa = 3.85PSKK317 pKa = 11.08PNAFPNYY324 pKa = 8.79MFAPDD329 pKa = 3.66SALSAVVTTDD339 pKa = 4.09SEE341 pKa = 4.87ADD343 pKa = 3.27TYY345 pKa = 10.98DD346 pKa = 3.58ARR348 pKa = 11.84RR349 pKa = 11.84MNYY352 pKa = 8.76MGRR355 pKa = 11.84TQEE358 pKa = 4.06AGQTRR363 pKa = 11.84TWYY366 pKa = 9.93QRR368 pKa = 11.84GVLMGGDD375 pKa = 3.57TDD377 pKa = 3.69ATTMSVYY384 pKa = 10.04MGEE387 pKa = 4.07AWLKK391 pKa = 11.26AEE393 pKa = 5.35LKK395 pKa = 10.64SQFLNMFNALPGITPDD411 pKa = 2.95IAGRR415 pKa = 11.84SYY417 pKa = 11.31INLYY421 pKa = 10.38LDD423 pKa = 3.67AAVAQALPDD432 pKa = 4.06GGNGMISVGKK442 pKa = 10.0QLTTTQQAYY451 pKa = 6.65ITQVSGDD458 pKa = 3.43EE459 pKa = 4.35DD460 pKa = 2.96AWKK463 pKa = 9.99QVQSEE468 pKa = 5.23GYY470 pKa = 8.11WYY472 pKa = 9.21TVNFFSTIAEE482 pKa = 4.18SGVTEE487 pKa = 4.22WTAEE491 pKa = 3.84YY492 pKa = 10.1TLIYY496 pKa = 9.42ATAEE500 pKa = 4.17KK501 pKa = 10.05VRR503 pKa = 11.84KK504 pKa = 9.94VSGTHH509 pKa = 5.91ILII512 pKa = 4.45
Molecular weight: 55.11 kDa
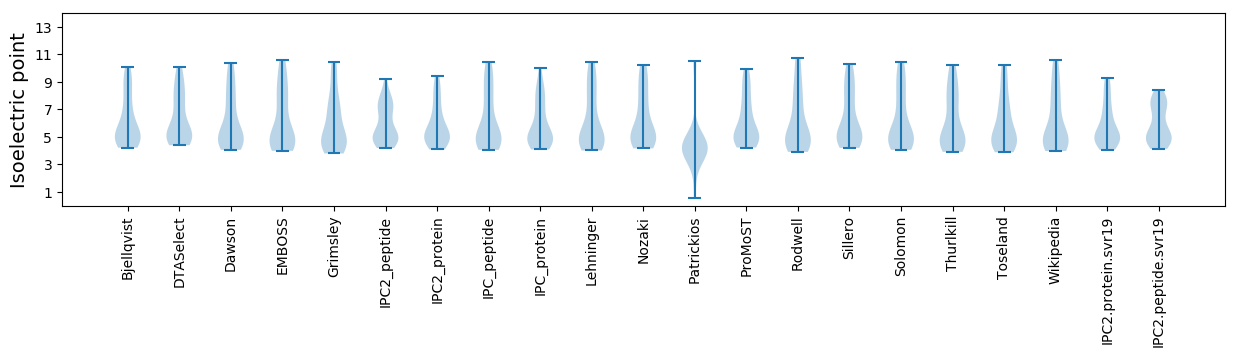
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6M3T8H9|A0A6M3T8H9_9CAUD Uncharacterized protein OS=Vibrio phage phiV039C OX=2705110 PE=4 SV=1
MM1 pKa = 6.94ITPRR5 pKa = 11.84QLQVLRR11 pKa = 11.84LYY13 pKa = 11.36ANGLTSIEE21 pKa = 4.07IAAEE25 pKa = 3.68LAISVRR31 pKa = 11.84TVEE34 pKa = 3.9EE35 pKa = 3.86HH36 pKa = 7.01LRR38 pKa = 11.84GCRR41 pKa = 11.84VEE43 pKa = 5.74LKK45 pKa = 10.64AKK47 pKa = 10.45NSVHH51 pKa = 6.26AVALAMQKK59 pKa = 10.86GIIKK63 pKa = 10.06LIILLSVYY71 pKa = 10.4QGMAADD77 pKa = 3.54YY78 pKa = 9.85HH79 pKa = 7.13EE80 pKa = 5.11DD81 pKa = 3.24MRR83 pKa = 11.84RR84 pKa = 11.84PPQVRR89 pKa = 11.84VRR91 pKa = 11.84ITRR94 pKa = 11.84TQRR97 pKa = 11.84RR98 pKa = 11.84EE99 pKa = 3.83CC100 pKa = 4.0
MM1 pKa = 6.94ITPRR5 pKa = 11.84QLQVLRR11 pKa = 11.84LYY13 pKa = 11.36ANGLTSIEE21 pKa = 4.07IAAEE25 pKa = 3.68LAISVRR31 pKa = 11.84TVEE34 pKa = 3.9EE35 pKa = 3.86HH36 pKa = 7.01LRR38 pKa = 11.84GCRR41 pKa = 11.84VEE43 pKa = 5.74LKK45 pKa = 10.64AKK47 pKa = 10.45NSVHH51 pKa = 6.26AVALAMQKK59 pKa = 10.86GIIKK63 pKa = 10.06LIILLSVYY71 pKa = 10.4QGMAADD77 pKa = 3.54YY78 pKa = 9.85HH79 pKa = 7.13EE80 pKa = 5.11DD81 pKa = 3.24MRR83 pKa = 11.84RR84 pKa = 11.84PPQVRR89 pKa = 11.84VRR91 pKa = 11.84ITRR94 pKa = 11.84TQRR97 pKa = 11.84RR98 pKa = 11.84EE99 pKa = 3.83CC100 pKa = 4.0
Molecular weight: 11.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
8952 |
72 |
812 |
248.7 |
27.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.764 ± 0.566 | 1.117 ± 0.167 |
6.49 ± 0.286 | 7.194 ± 0.656 |
3.943 ± 0.271 | 7.149 ± 0.396 |
1.597 ± 0.236 | 6.133 ± 0.282 |
6.591 ± 0.521 | 7.373 ± 0.289 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.659 ± 0.19 | 5.496 ± 0.281 |
3.664 ± 0.267 | 4.144 ± 0.367 |
4.457 ± 0.386 | 7.283 ± 0.534 |
5.831 ± 0.434 | 6.311 ± 0.343 |
1.408 ± 0.15 | 3.396 ± 0.319 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
