
Planctomycetes bacterium HG15A2
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Planctomycetia; Pirellulales; Lacipirellulaceae; Adhaeretor
Average proteome isoelectric point is 6.01
Get precalculated fractions of proteins
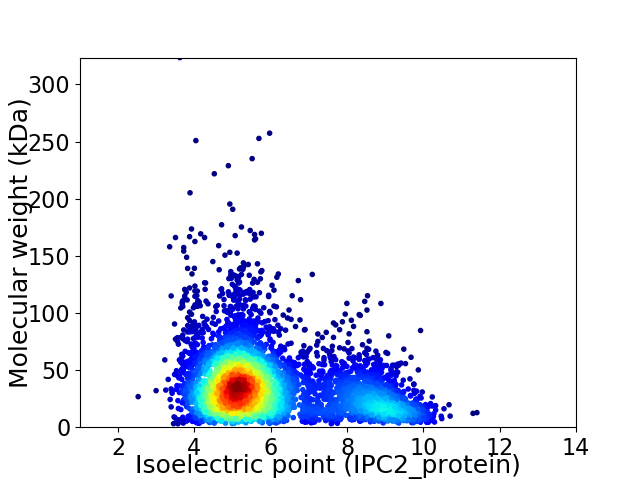
Virtual 2D-PAGE plot for 4864 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
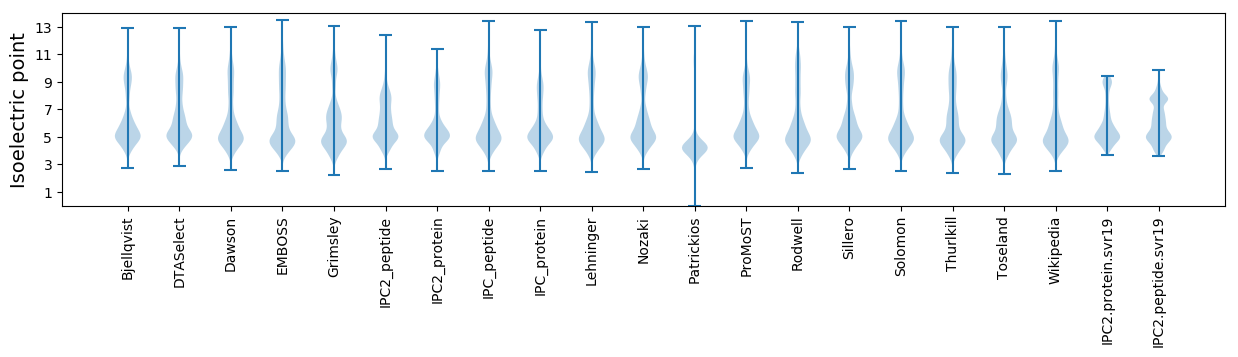
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A517MU95|A0A517MU95_9BACT Phosphorylated carbohydrates phosphatase OS=Planctomycetes bacterium HG15A2 OX=1930276 GN=HG15A2_17280 PE=4 SV=1
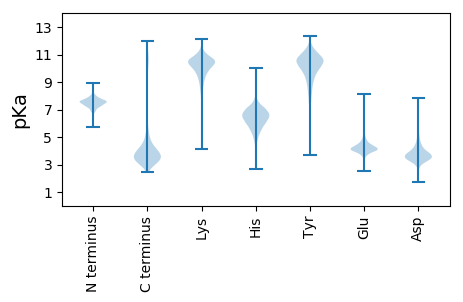
MM1 pKa = 6.86NCFYY5 pKa = 10.75IFQTLFNSFKK15 pKa = 9.96GSLRR19 pKa = 11.84FVVAKK24 pKa = 10.25VPLRR28 pKa = 11.84ACVAAAVLADD38 pKa = 3.95MACSASAITIADD50 pKa = 3.43AAGDD54 pKa = 3.83YY55 pKa = 10.43LAAAGSTSTAPLSPPTGWSYY75 pKa = 11.63LGADD79 pKa = 3.93MPNGGTEE86 pKa = 3.75ISLTAGDD93 pKa = 4.36VGNQGASYY101 pKa = 10.02QGFVGGSTAGTAAVYY116 pKa = 7.61GTNTASSLEE125 pKa = 3.91FEE127 pKa = 4.4IFSNGEE133 pKa = 3.7DD134 pKa = 3.16NGAVVGTDD142 pKa = 3.62LLLHH146 pKa = 6.89PGQGGNADD154 pKa = 3.58EE155 pKa = 4.65FVIARR160 pKa = 11.84YY161 pKa = 8.03TISTADD167 pKa = 3.51AANAAAGTGSISGSFRR183 pKa = 11.84EE184 pKa = 4.5LIIGGGAAVDD194 pKa = 4.49SISADD199 pKa = 2.8IYY201 pKa = 11.2HH202 pKa = 5.95NTNNLFSVTGNQSTANALTQAEE224 pKa = 4.87GTFNLTGLTFAEE236 pKa = 4.4NDD238 pKa = 3.79TIDD241 pKa = 3.86FVVGINGHH249 pKa = 6.35FGADD253 pKa = 3.35EE254 pKa = 4.04TALQAIIEE262 pKa = 4.35VEE264 pKa = 4.11QTQPTEE270 pKa = 3.83LLTLQVHH277 pKa = 5.57TLSGDD282 pKa = 3.12VSLINGSQPQDD293 pKa = 2.59IDD295 pKa = 4.11FYY297 pKa = 11.24SIASLGVALNFDD309 pKa = 4.22GWNSLEE315 pKa = 4.34DD316 pKa = 3.54QDD318 pKa = 5.92FEE320 pKa = 5.62GNGAPGTGNGWEE332 pKa = 4.0QSGGSGDD339 pKa = 3.8TVLTEE344 pKa = 4.52AYY346 pKa = 7.99LTGSSVIAADD356 pKa = 3.62SSISIGSAFDD366 pKa = 3.39PTEE369 pKa = 4.3FGAGNDD375 pKa = 3.5GDD377 pKa = 4.52LTFQYY382 pKa = 10.74RR383 pKa = 11.84LADD386 pKa = 3.75GSVVPGLVEE395 pKa = 4.16YY396 pKa = 8.97VTSGDD401 pKa = 3.37FDD403 pKa = 5.89ADD405 pKa = 3.81LDD407 pKa = 3.85IDD409 pKa = 4.42GVDD412 pKa = 4.1FLAWQRR418 pKa = 11.84GFGTMFDD425 pKa = 3.95ASDD428 pKa = 3.79LEE430 pKa = 4.31DD431 pKa = 3.53WQDD434 pKa = 3.02HH435 pKa = 5.77YY436 pKa = 11.84GAGGTSPSLAVAVPEE451 pKa = 4.03PASIVLVIFSLLAGQGFFRR470 pKa = 11.84RR471 pKa = 11.84QSRR474 pKa = 11.84PARR477 pKa = 3.78
MM1 pKa = 6.86NCFYY5 pKa = 10.75IFQTLFNSFKK15 pKa = 9.96GSLRR19 pKa = 11.84FVVAKK24 pKa = 10.25VPLRR28 pKa = 11.84ACVAAAVLADD38 pKa = 3.95MACSASAITIADD50 pKa = 3.43AAGDD54 pKa = 3.83YY55 pKa = 10.43LAAAGSTSTAPLSPPTGWSYY75 pKa = 11.63LGADD79 pKa = 3.93MPNGGTEE86 pKa = 3.75ISLTAGDD93 pKa = 4.36VGNQGASYY101 pKa = 10.02QGFVGGSTAGTAAVYY116 pKa = 7.61GTNTASSLEE125 pKa = 3.91FEE127 pKa = 4.4IFSNGEE133 pKa = 3.7DD134 pKa = 3.16NGAVVGTDD142 pKa = 3.62LLLHH146 pKa = 6.89PGQGGNADD154 pKa = 3.58EE155 pKa = 4.65FVIARR160 pKa = 11.84YY161 pKa = 8.03TISTADD167 pKa = 3.51AANAAAGTGSISGSFRR183 pKa = 11.84EE184 pKa = 4.5LIIGGGAAVDD194 pKa = 4.49SISADD199 pKa = 2.8IYY201 pKa = 11.2HH202 pKa = 5.95NTNNLFSVTGNQSTANALTQAEE224 pKa = 4.87GTFNLTGLTFAEE236 pKa = 4.4NDD238 pKa = 3.79TIDD241 pKa = 3.86FVVGINGHH249 pKa = 6.35FGADD253 pKa = 3.35EE254 pKa = 4.04TALQAIIEE262 pKa = 4.35VEE264 pKa = 4.11QTQPTEE270 pKa = 3.83LLTLQVHH277 pKa = 5.57TLSGDD282 pKa = 3.12VSLINGSQPQDD293 pKa = 2.59IDD295 pKa = 4.11FYY297 pKa = 11.24SIASLGVALNFDD309 pKa = 4.22GWNSLEE315 pKa = 4.34DD316 pKa = 3.54QDD318 pKa = 5.92FEE320 pKa = 5.62GNGAPGTGNGWEE332 pKa = 4.0QSGGSGDD339 pKa = 3.8TVLTEE344 pKa = 4.52AYY346 pKa = 7.99LTGSSVIAADD356 pKa = 3.62SSISIGSAFDD366 pKa = 3.39PTEE369 pKa = 4.3FGAGNDD375 pKa = 3.5GDD377 pKa = 4.52LTFQYY382 pKa = 10.74RR383 pKa = 11.84LADD386 pKa = 3.75GSVVPGLVEE395 pKa = 4.16YY396 pKa = 8.97VTSGDD401 pKa = 3.37FDD403 pKa = 5.89ADD405 pKa = 3.81LDD407 pKa = 3.85IDD409 pKa = 4.42GVDD412 pKa = 4.1FLAWQRR418 pKa = 11.84GFGTMFDD425 pKa = 3.95ASDD428 pKa = 3.79LEE430 pKa = 4.31DD431 pKa = 3.53WQDD434 pKa = 3.02HH435 pKa = 5.77YY436 pKa = 11.84GAGGTSPSLAVAVPEE451 pKa = 4.03PASIVLVIFSLLAGQGFFRR470 pKa = 11.84RR471 pKa = 11.84QSRR474 pKa = 11.84PARR477 pKa = 3.78
Molecular weight: 49.03 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A517N2B3|A0A517N2B3_9BACT Uncharacterized protein OS=Planctomycetes bacterium HG15A2 OX=1930276 GN=HG15A2_46030 PE=4 SV=1
MM1 pKa = 7.47AAAKK5 pKa = 8.44KK6 pKa = 6.88TAKK9 pKa = 10.27RR10 pKa = 11.84GAAKK14 pKa = 8.91TAKK17 pKa = 10.07KK18 pKa = 10.25SPKK21 pKa = 9.63KK22 pKa = 10.1RR23 pKa = 11.84ATAKK27 pKa = 10.44KK28 pKa = 8.33STAKK32 pKa = 10.42KK33 pKa = 9.78SPKK36 pKa = 9.46KK37 pKa = 10.02RR38 pKa = 11.84AAKK41 pKa = 10.17KK42 pKa = 9.25ATTKK46 pKa = 10.44KK47 pKa = 10.17AAKK50 pKa = 9.79KK51 pKa = 9.97SPKK54 pKa = 10.0KK55 pKa = 10.34KK56 pKa = 8.83ATKK59 pKa = 9.92KK60 pKa = 9.69AAKK63 pKa = 9.85KK64 pKa = 8.79STAKK68 pKa = 9.62RR69 pKa = 11.84TTKK72 pKa = 10.4KK73 pKa = 10.57SPKK76 pKa = 9.38KK77 pKa = 9.53KK78 pKa = 9.86AAAKK82 pKa = 7.94RR83 pKa = 11.84TTKK86 pKa = 10.54KK87 pKa = 9.4STAKK91 pKa = 10.39KK92 pKa = 8.42STAKK96 pKa = 10.36RR97 pKa = 11.84SPKK100 pKa = 9.03KK101 pKa = 7.25TTAKK105 pKa = 10.12RR106 pKa = 11.84GRR108 pKa = 11.84PAKK111 pKa = 10.29KK112 pKa = 9.89KK113 pKa = 10.37
MM1 pKa = 7.47AAAKK5 pKa = 8.44KK6 pKa = 6.88TAKK9 pKa = 10.27RR10 pKa = 11.84GAAKK14 pKa = 8.91TAKK17 pKa = 10.07KK18 pKa = 10.25SPKK21 pKa = 9.63KK22 pKa = 10.1RR23 pKa = 11.84ATAKK27 pKa = 10.44KK28 pKa = 8.33STAKK32 pKa = 10.42KK33 pKa = 9.78SPKK36 pKa = 9.46KK37 pKa = 10.02RR38 pKa = 11.84AAKK41 pKa = 10.17KK42 pKa = 9.25ATTKK46 pKa = 10.44KK47 pKa = 10.17AAKK50 pKa = 9.79KK51 pKa = 9.97SPKK54 pKa = 10.0KK55 pKa = 10.34KK56 pKa = 8.83ATKK59 pKa = 9.92KK60 pKa = 9.69AAKK63 pKa = 9.85KK64 pKa = 8.79STAKK68 pKa = 9.62RR69 pKa = 11.84TTKK72 pKa = 10.4KK73 pKa = 10.57SPKK76 pKa = 9.38KK77 pKa = 9.53KK78 pKa = 9.86AAAKK82 pKa = 7.94RR83 pKa = 11.84TTKK86 pKa = 10.54KK87 pKa = 9.4STAKK91 pKa = 10.39KK92 pKa = 8.42STAKK96 pKa = 10.36RR97 pKa = 11.84SPKK100 pKa = 9.03KK101 pKa = 7.25TTAKK105 pKa = 10.12RR106 pKa = 11.84GRR108 pKa = 11.84PAKK111 pKa = 10.29KK112 pKa = 9.89KK113 pKa = 10.37
Molecular weight: 12.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1702111 |
29 |
3026 |
349.9 |
38.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.807 ± 0.032 | 1.106 ± 0.014 |
5.896 ± 0.035 | 6.521 ± 0.038 |
3.681 ± 0.026 | 8.073 ± 0.041 |
2.104 ± 0.017 | 4.818 ± 0.026 |
3.769 ± 0.032 | 10.083 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.075 ± 0.017 | 3.446 ± 0.029 |
4.949 ± 0.027 | 4.108 ± 0.026 |
5.99 ± 0.032 | 6.68 ± 0.034 |
5.785 ± 0.031 | 7.123 ± 0.032 |
1.477 ± 0.014 | 2.51 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
