
Candidatus Electrothrix aarhusiensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfobacterales; Desulfobulbaceae; Candidatus Electrothrix
Average proteome isoelectric point is 6.62
Get precalculated fractions of proteins
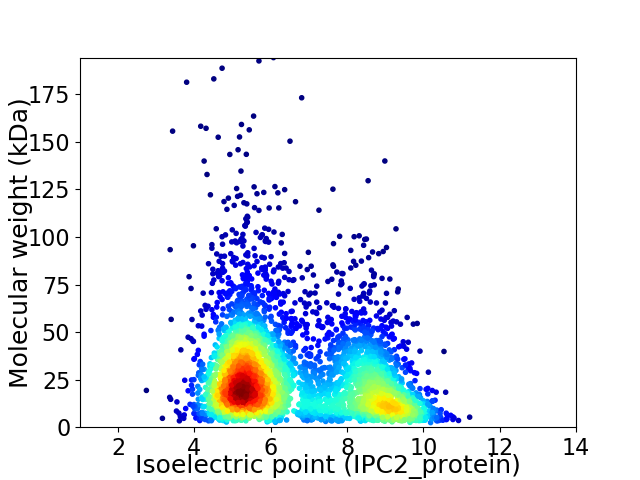
Virtual 2D-PAGE plot for 4071 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A444ISR4|A0A444ISR4_9DELT PD-(D/E)XK nuclease superfamily protein OS=Candidatus Electrothrix aarhusiensis OX=1859131 GN=H206_02607 PE=4 SV=1
MM1 pKa = 8.19PDD3 pKa = 2.94RR4 pKa = 11.84TFGDD8 pKa = 3.33KK9 pKa = 10.87GVLITDD15 pKa = 4.52DD16 pKa = 5.3DD17 pKa = 4.92NDD19 pKa = 4.06TVFYY23 pKa = 10.8DD24 pKa = 3.54VLMTEE29 pKa = 4.45KK30 pKa = 9.83MIAATGVTVGEE41 pKa = 4.2EE42 pKa = 4.2EE43 pKa = 4.0MRR45 pKa = 11.84EE46 pKa = 4.02ALLITYY52 pKa = 8.9AQGDD56 pKa = 4.12GANSVQLFQQQVAAASEE73 pKa = 4.41GAAEE77 pKa = 4.08ATEE80 pKa = 4.38AADD83 pKa = 3.79EE84 pKa = 4.56DD85 pKa = 4.37SEE87 pKa = 4.35EE88 pKa = 4.51TEE90 pKa = 4.21TVAQIVTTEE99 pKa = 3.57VDD101 pKa = 3.23NEE103 pKa = 3.51EE104 pKa = 5.14DD105 pKa = 4.0YY106 pKa = 11.76AFSLAAADD114 pKa = 3.83AGSVVVVGASGAQEE128 pKa = 3.69IASAAVRR135 pKa = 11.84KK136 pKa = 7.8YY137 pKa = 9.38TIFQSSVTGSSWSTATGNASILTGDD162 pKa = 3.72AQNVTRR168 pKa = 11.84TTALIPGEE176 pKa = 4.18VLSGLGTVTEE186 pKa = 4.25RR187 pKa = 11.84GVVFSTDD194 pKa = 3.58PLPVLKK200 pKa = 10.78GSDD203 pKa = 3.46STTDD207 pKa = 3.29DD208 pKa = 3.69TTDD211 pKa = 3.44DD212 pKa = 3.96TTDD215 pKa = 3.49DD216 pKa = 3.85TTTDD220 pKa = 3.85DD221 pKa = 4.52DD222 pKa = 4.14TDD224 pKa = 3.6EE225 pKa = 4.62TGPSITSSTEE235 pKa = 3.69SSFLTTEE242 pKa = 3.76QVILVVSTDD251 pKa = 3.18EE252 pKa = 4.78NATCRR257 pKa = 11.84YY258 pKa = 7.88NVKK261 pKa = 10.29YY262 pKa = 10.84DD263 pKa = 3.56EE264 pKa = 5.97DD265 pKa = 4.09YY266 pKa = 11.86GEE268 pKa = 4.19MGGILSSSFGTGHH281 pKa = 6.95HH282 pKa = 6.29IVLGTLTEE290 pKa = 4.09GDD292 pKa = 3.44YY293 pKa = 11.01TFYY296 pKa = 11.07VRR298 pKa = 11.84CKK300 pKa = 10.42DD301 pKa = 3.34EE302 pKa = 4.45SGNISTAGTEE312 pKa = 3.88VTFAVSADD320 pKa = 3.57DD321 pKa = 3.01TGAPVRR327 pKa = 11.84SEE329 pKa = 4.55LSPSSTVSSTSVTLSLSTDD348 pKa = 3.01VAATCKK354 pKa = 9.76YY355 pKa = 5.36TTSDD359 pKa = 3.57SGMDD363 pKa = 3.54YY364 pKa = 11.61NDD366 pKa = 3.33MKK368 pKa = 11.2KK369 pKa = 10.34EE370 pKa = 3.74FDD372 pKa = 3.25ATGGISHH379 pKa = 6.45SEE381 pKa = 4.27SVSSLVVGEE390 pKa = 4.42SYY392 pKa = 10.3TYY394 pKa = 10.08YY395 pKa = 10.52VRR397 pKa = 11.84CKK399 pKa = 10.54SISTGVANTSSTEE412 pKa = 3.55ISFTVDD418 pKa = 3.32DD419 pKa = 4.58TDD421 pKa = 3.69STAPTITSTTSSTFAGTDD439 pKa = 3.48EE440 pKa = 4.86VILTVTTNEE449 pKa = 3.9TAYY452 pKa = 10.48CGYY455 pKa = 10.47SDD457 pKa = 4.73VYY459 pKa = 10.71SIDD462 pKa = 3.42MADD465 pKa = 3.67YY466 pKa = 8.77EE467 pKa = 4.79ANPMVTSDD475 pKa = 3.67GVTHH479 pKa = 6.84KK480 pKa = 10.97ASLGVLSTASYY491 pKa = 9.32TYY493 pKa = 10.94YY494 pKa = 10.0IGCYY498 pKa = 9.71DD499 pKa = 3.41EE500 pKa = 5.17SGNVTTFPAEE510 pKa = 3.65ILFDD514 pKa = 3.71VASLYY519 pKa = 10.99KK520 pKa = 10.71NSIFYY525 pKa = 10.95AEE527 pKa = 4.19NQVASSSPMKK537 pKa = 10.75AGFTTALEE545 pKa = 4.39SVGNLFVSTAIAQDD559 pKa = 3.6TTDD562 pKa = 3.82TTDD565 pKa = 2.87STTDD569 pKa = 3.02TTDD572 pKa = 3.88ADD574 pKa = 4.2TEE576 pKa = 4.19DD577 pKa = 4.2SNFLEE582 pKa = 4.75EE583 pKa = 4.48GSVDD587 pKa = 3.49EE588 pKa = 5.47GSGTGQFTSKK598 pKa = 11.13LEE600 pKa = 4.01DD601 pKa = 3.61LKK603 pKa = 11.22PGTFFYY609 pKa = 11.1ARR611 pKa = 11.84AYY613 pKa = 10.31AVIGGTTYY621 pKa = 10.97YY622 pKa = 10.9GNQISFQTADD632 pKa = 3.32SCFVATAAYY641 pKa = 10.02GSLFHH646 pKa = 7.09PAVQLLRR653 pKa = 11.84DD654 pKa = 3.59FRR656 pKa = 11.84DD657 pKa = 3.25RR658 pKa = 11.84FMLDD662 pKa = 3.18NPVSRR667 pKa = 11.84SMVRR671 pKa = 11.84LYY673 pKa = 10.95YY674 pKa = 10.24RR675 pKa = 11.84YY676 pKa = 9.76SPPIADD682 pKa = 4.02VISSNTVLRR691 pKa = 11.84PVTRR695 pKa = 11.84TLLLPIVGSAWLTMHH710 pKa = 7.15FGWLWLILPAATMALLSWFGMQRR733 pKa = 11.84MQAARR738 pKa = 11.84KK739 pKa = 9.57EE740 pKa = 4.17EE741 pKa = 4.0LAA743 pKa = 4.54
MM1 pKa = 8.19PDD3 pKa = 2.94RR4 pKa = 11.84TFGDD8 pKa = 3.33KK9 pKa = 10.87GVLITDD15 pKa = 4.52DD16 pKa = 5.3DD17 pKa = 4.92NDD19 pKa = 4.06TVFYY23 pKa = 10.8DD24 pKa = 3.54VLMTEE29 pKa = 4.45KK30 pKa = 9.83MIAATGVTVGEE41 pKa = 4.2EE42 pKa = 4.2EE43 pKa = 4.0MRR45 pKa = 11.84EE46 pKa = 4.02ALLITYY52 pKa = 8.9AQGDD56 pKa = 4.12GANSVQLFQQQVAAASEE73 pKa = 4.41GAAEE77 pKa = 4.08ATEE80 pKa = 4.38AADD83 pKa = 3.79EE84 pKa = 4.56DD85 pKa = 4.37SEE87 pKa = 4.35EE88 pKa = 4.51TEE90 pKa = 4.21TVAQIVTTEE99 pKa = 3.57VDD101 pKa = 3.23NEE103 pKa = 3.51EE104 pKa = 5.14DD105 pKa = 4.0YY106 pKa = 11.76AFSLAAADD114 pKa = 3.83AGSVVVVGASGAQEE128 pKa = 3.69IASAAVRR135 pKa = 11.84KK136 pKa = 7.8YY137 pKa = 9.38TIFQSSVTGSSWSTATGNASILTGDD162 pKa = 3.72AQNVTRR168 pKa = 11.84TTALIPGEE176 pKa = 4.18VLSGLGTVTEE186 pKa = 4.25RR187 pKa = 11.84GVVFSTDD194 pKa = 3.58PLPVLKK200 pKa = 10.78GSDD203 pKa = 3.46STTDD207 pKa = 3.29DD208 pKa = 3.69TTDD211 pKa = 3.44DD212 pKa = 3.96TTDD215 pKa = 3.49DD216 pKa = 3.85TTTDD220 pKa = 3.85DD221 pKa = 4.52DD222 pKa = 4.14TDD224 pKa = 3.6EE225 pKa = 4.62TGPSITSSTEE235 pKa = 3.69SSFLTTEE242 pKa = 3.76QVILVVSTDD251 pKa = 3.18EE252 pKa = 4.78NATCRR257 pKa = 11.84YY258 pKa = 7.88NVKK261 pKa = 10.29YY262 pKa = 10.84DD263 pKa = 3.56EE264 pKa = 5.97DD265 pKa = 4.09YY266 pKa = 11.86GEE268 pKa = 4.19MGGILSSSFGTGHH281 pKa = 6.95HH282 pKa = 6.29IVLGTLTEE290 pKa = 4.09GDD292 pKa = 3.44YY293 pKa = 11.01TFYY296 pKa = 11.07VRR298 pKa = 11.84CKK300 pKa = 10.42DD301 pKa = 3.34EE302 pKa = 4.45SGNISTAGTEE312 pKa = 3.88VTFAVSADD320 pKa = 3.57DD321 pKa = 3.01TGAPVRR327 pKa = 11.84SEE329 pKa = 4.55LSPSSTVSSTSVTLSLSTDD348 pKa = 3.01VAATCKK354 pKa = 9.76YY355 pKa = 5.36TTSDD359 pKa = 3.57SGMDD363 pKa = 3.54YY364 pKa = 11.61NDD366 pKa = 3.33MKK368 pKa = 11.2KK369 pKa = 10.34EE370 pKa = 3.74FDD372 pKa = 3.25ATGGISHH379 pKa = 6.45SEE381 pKa = 4.27SVSSLVVGEE390 pKa = 4.42SYY392 pKa = 10.3TYY394 pKa = 10.08YY395 pKa = 10.52VRR397 pKa = 11.84CKK399 pKa = 10.54SISTGVANTSSTEE412 pKa = 3.55ISFTVDD418 pKa = 3.32DD419 pKa = 4.58TDD421 pKa = 3.69STAPTITSTTSSTFAGTDD439 pKa = 3.48EE440 pKa = 4.86VILTVTTNEE449 pKa = 3.9TAYY452 pKa = 10.48CGYY455 pKa = 10.47SDD457 pKa = 4.73VYY459 pKa = 10.71SIDD462 pKa = 3.42MADD465 pKa = 3.67YY466 pKa = 8.77EE467 pKa = 4.79ANPMVTSDD475 pKa = 3.67GVTHH479 pKa = 6.84KK480 pKa = 10.97ASLGVLSTASYY491 pKa = 9.32TYY493 pKa = 10.94YY494 pKa = 10.0IGCYY498 pKa = 9.71DD499 pKa = 3.41EE500 pKa = 5.17SGNVTTFPAEE510 pKa = 3.65ILFDD514 pKa = 3.71VASLYY519 pKa = 10.99KK520 pKa = 10.71NSIFYY525 pKa = 10.95AEE527 pKa = 4.19NQVASSSPMKK537 pKa = 10.75AGFTTALEE545 pKa = 4.39SVGNLFVSTAIAQDD559 pKa = 3.6TTDD562 pKa = 3.82TTDD565 pKa = 2.87STTDD569 pKa = 3.02TTDD572 pKa = 3.88ADD574 pKa = 4.2TEE576 pKa = 4.19DD577 pKa = 4.2SNFLEE582 pKa = 4.75EE583 pKa = 4.48GSVDD587 pKa = 3.49EE588 pKa = 5.47GSGTGQFTSKK598 pKa = 11.13LEE600 pKa = 4.01DD601 pKa = 3.61LKK603 pKa = 11.22PGTFFYY609 pKa = 11.1ARR611 pKa = 11.84AYY613 pKa = 10.31AVIGGTTYY621 pKa = 10.97YY622 pKa = 10.9GNQISFQTADD632 pKa = 3.32SCFVATAAYY641 pKa = 10.02GSLFHH646 pKa = 7.09PAVQLLRR653 pKa = 11.84DD654 pKa = 3.59FRR656 pKa = 11.84DD657 pKa = 3.25RR658 pKa = 11.84FMLDD662 pKa = 3.18NPVSRR667 pKa = 11.84SMVRR671 pKa = 11.84LYY673 pKa = 10.95YY674 pKa = 10.24RR675 pKa = 11.84YY676 pKa = 9.76SPPIADD682 pKa = 4.02VISSNTVLRR691 pKa = 11.84PVTRR695 pKa = 11.84TLLLPIVGSAWLTMHH710 pKa = 7.15FGWLWLILPAATMALLSWFGMQRR733 pKa = 11.84MQAARR738 pKa = 11.84KK739 pKa = 9.57EE740 pKa = 4.17EE741 pKa = 4.0LAA743 pKa = 4.54
Molecular weight: 79.13 kDa
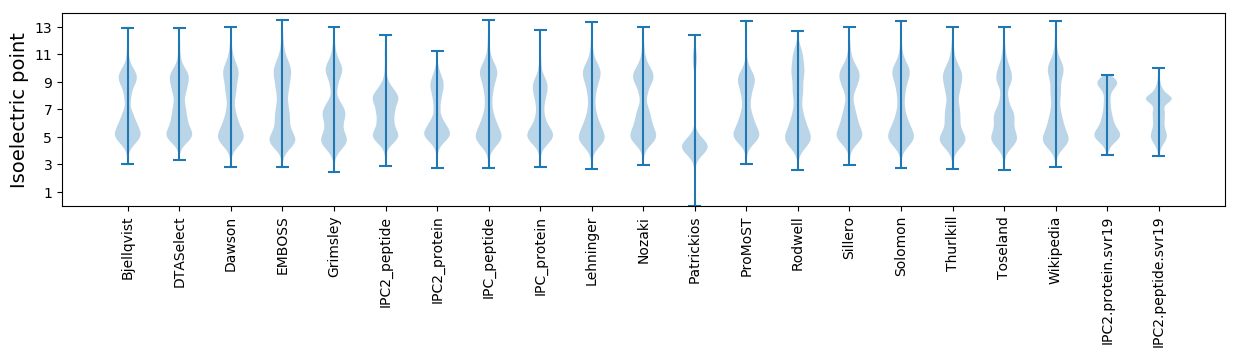
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S3QQZ8|A0A3S3QQZ8_9DELT Uncharacterized protein OS=Candidatus Electrothrix aarhusiensis OX=1859131 GN=H206_01075 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNLKK12 pKa = 10.14RR13 pKa = 11.84KK14 pKa = 8.66RR15 pKa = 11.84SHH17 pKa = 5.99GFRR20 pKa = 11.84VRR22 pKa = 11.84MKK24 pKa = 9.0TASGQAIIKK33 pKa = 9.81RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.88GRR40 pKa = 11.84KK41 pKa = 8.85RR42 pKa = 11.84LSAA45 pKa = 3.96
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNLKK12 pKa = 10.14RR13 pKa = 11.84KK14 pKa = 8.66RR15 pKa = 11.84SHH17 pKa = 5.99GFRR20 pKa = 11.84VRR22 pKa = 11.84MKK24 pKa = 9.0TASGQAIIKK33 pKa = 9.81RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.88GRR40 pKa = 11.84KK41 pKa = 8.85RR42 pKa = 11.84LSAA45 pKa = 3.96
Molecular weight: 5.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1082977 |
21 |
1712 |
266.0 |
29.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.926 ± 0.04 | 1.389 ± 0.019 |
5.455 ± 0.043 | 6.806 ± 0.042 |
4.358 ± 0.031 | 6.973 ± 0.042 |
2.146 ± 0.018 | 6.485 ± 0.036 |
5.837 ± 0.044 | 10.425 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.496 ± 0.02 | 3.795 ± 0.032 |
4.277 ± 0.027 | 4.199 ± 0.031 |
5.424 ± 0.029 | 6.183 ± 0.034 |
5.215 ± 0.034 | 6.44 ± 0.033 |
1.14 ± 0.018 | 3.029 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
