
Solemya velum gill symbiont
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Gammaproteobacteria incertae sedis; sulfur-oxidizing symbionts
Average proteome isoelectric point is 5.96
Get precalculated fractions of proteins
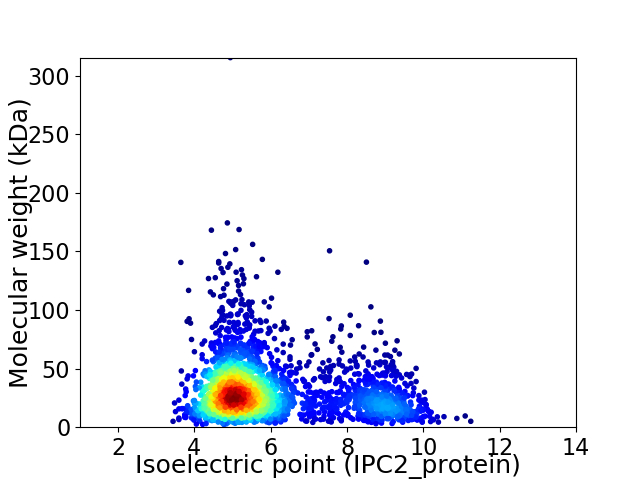
Virtual 2D-PAGE plot for 2657 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B0H5F3|A0A0B0H5F3_SOVGS 50S ribosomal protein L15 OS=Solemya velum gill symbiont OX=2340 GN=rplO PE=3 SV=1
MM1 pKa = 6.88NHH3 pKa = 6.88KK4 pKa = 10.7ASLNQNKK11 pKa = 9.62SVEE14 pKa = 4.26PNSLQGMRR22 pKa = 11.84RR23 pKa = 11.84TATAIAVASVLSVATLPQATAGFSYY48 pKa = 11.02GSGDD52 pKa = 3.69TIAITGPVTAHH63 pKa = 6.29NNDD66 pKa = 2.9GSDD69 pKa = 3.46LVNVNAYY76 pKa = 9.68FADD79 pKa = 5.4DD80 pKa = 3.21ISGSYY85 pKa = 10.48GVLLRR90 pKa = 11.84GDD92 pKa = 4.08SGASSNTITNNGYY105 pKa = 9.93DD106 pKa = 3.72ADD108 pKa = 4.16VSGSAAAIEE117 pKa = 4.41VGYY120 pKa = 10.46IPGPDD125 pKa = 3.48DD126 pKa = 4.1VRR128 pKa = 11.84AHH130 pKa = 6.54NNTITNSGRR139 pKa = 11.84MEE141 pKa = 4.4GSGGRR146 pKa = 11.84AVPTGRR152 pKa = 11.84TGVAPRR158 pKa = 11.84IVITPPGTYY167 pKa = 9.45LQYY170 pKa = 10.85EE171 pKa = 4.63GTVHH175 pKa = 5.58ITAFAAEE182 pKa = 4.41NNTDD186 pKa = 3.39ADD188 pKa = 3.97AEE190 pKa = 4.22VDD192 pKa = 3.54NNLIEE197 pKa = 4.22NSGTISGDD205 pKa = 2.8GTLAVHH211 pKa = 6.82LGSFAYY217 pKa = 10.59VSGSSAHH224 pKa = 6.86KK225 pKa = 9.48GTAEE229 pKa = 3.84VGNNSVNNSGGIVSDD244 pKa = 3.54STGVQFLSIAGGFSGTHH261 pKa = 6.41AEE263 pKa = 4.03ATVNDD268 pKa = 3.85NVVSNAYY275 pKa = 10.38DD276 pKa = 3.5GDD278 pKa = 4.1INGGMLSGIDD288 pKa = 3.5MTAIAATSSSCCGPQGNADD307 pKa = 3.44ASVEE311 pKa = 4.38GNSNTNDD318 pKa = 3.3GYY320 pKa = 11.53AGDD323 pKa = 4.48SLLSGASMNAIAASGYY339 pKa = 9.48MPFARR344 pKa = 11.84ADD346 pKa = 3.49AEE348 pKa = 4.57VNSNEE353 pKa = 3.65ITNAEE358 pKa = 3.98NANIEE363 pKa = 4.15GKK365 pKa = 10.87YY366 pKa = 10.64NGIALRR372 pKa = 11.84AAGEE376 pKa = 4.29DD377 pKa = 2.8IKK379 pKa = 11.43YY380 pKa = 10.78AGDD383 pKa = 3.46VAVNSNMISNDD394 pKa = 2.81GTVFASEE401 pKa = 4.36DD402 pKa = 3.46QFADD406 pKa = 3.64SAVKK410 pKa = 9.98LASYY414 pKa = 11.08GNLANMFSKK423 pKa = 10.77DD424 pKa = 4.13DD425 pKa = 3.6GHH427 pKa = 6.74GLPFANDD434 pKa = 3.05TDD436 pKa = 4.35VNSNTITNSSTGDD449 pKa = 3.52MVNSAWMVRR458 pKa = 11.84DD459 pKa = 3.39SAVSLRR465 pKa = 11.84ADD467 pKa = 3.07SSTGDD472 pKa = 3.24RR473 pKa = 11.84YY474 pKa = 9.82DD475 pKa = 3.54TPFSGSSSAEE485 pKa = 3.88VNSNSIEE492 pKa = 3.77NHH494 pKa = 4.91GFIGSFAQYY503 pKa = 10.77NFGGGIEE510 pKa = 4.11LTADD514 pKa = 3.38AGGDD518 pKa = 3.36AYY520 pKa = 10.12EE521 pKa = 4.12YY522 pKa = 10.67DD523 pKa = 4.41YY524 pKa = 11.21ISSGGFIDD532 pKa = 3.4HH533 pKa = 6.75EE534 pKa = 4.25EE535 pKa = 4.41RR536 pKa = 11.84IGVSAGTADD545 pKa = 3.6VNKK548 pKa = 8.84NTVSNTSSDD557 pKa = 3.75TIAVGGYY564 pKa = 8.99VLFDD568 pKa = 3.82ADD570 pKa = 5.01GIEE573 pKa = 4.33LNADD577 pKa = 3.29ASASAEE583 pKa = 4.19TYY585 pKa = 10.33IDD587 pKa = 3.2ISGGFSDD594 pKa = 5.07ASGSADD600 pKa = 3.08SLAFAGDD607 pKa = 3.31ADD609 pKa = 4.05VNNNQVEE616 pKa = 4.27NSGSIEE622 pKa = 3.76VTGYY626 pKa = 9.55MGEE629 pKa = 4.15DD630 pKa = 3.14SQGIVLAADD639 pKa = 3.44AWAYY643 pKa = 10.87ANSDD647 pKa = 3.12VTAQSSGSSSADD659 pKa = 3.23SFGMASGGTTFVNNNDD675 pKa = 3.92VINSGDD681 pKa = 3.41ILTSGEE687 pKa = 4.56DD688 pKa = 3.66FPSQYY693 pKa = 9.34MQGILLTSSASAFAYY708 pKa = 10.53SEE710 pKa = 4.04ATAVSGGGNGDD721 pKa = 3.43ASGGATAMAGTAEE734 pKa = 4.03VHH736 pKa = 6.16NNNVVNSGTISTDD749 pKa = 3.33GEE751 pKa = 4.19DD752 pKa = 3.23SSGIEE757 pKa = 3.68LSAFAWADD765 pKa = 3.04ADD767 pKa = 3.72VDD769 pKa = 4.3LSGSSTTDD777 pKa = 2.58SDD779 pKa = 3.61GTTASGGFASVANNTITNSGDD800 pKa = 2.55ITTAGGRR807 pKa = 11.84GASSTGILLYY817 pKa = 10.85ADD819 pKa = 3.66VSAWAEE825 pKa = 4.03RR826 pKa = 11.84FSDD829 pKa = 3.84DD830 pKa = 3.95TDD832 pKa = 4.16GSDD835 pKa = 3.19VARR838 pKa = 11.84AGIAEE843 pKa = 4.06ITGNSVDD850 pKa = 3.44NSGVIDD856 pKa = 3.44AWEE859 pKa = 4.97GIVLEE864 pKa = 4.47ARR866 pKa = 11.84ASAFADD872 pKa = 3.22NYY874 pKa = 10.31ARR876 pKa = 11.84SGEE879 pKa = 4.23AYY881 pKa = 9.24LTDD884 pKa = 3.46NTITNSGTIIADD896 pKa = 3.4NVGIAIRR903 pKa = 11.84GEE905 pKa = 3.92ASGSVINTIFTHH917 pKa = 6.34GNSVDD922 pKa = 3.41NSGSMEE928 pKa = 4.14VYY930 pKa = 9.6NGSGIIVGEE939 pKa = 4.11EE940 pKa = 4.03APSGSGSYY948 pKa = 8.53TWDD951 pKa = 3.08NTINNTGTILAGEE964 pKa = 4.21DD965 pKa = 3.53TAIGIYY971 pKa = 10.53GDD973 pKa = 3.45AEE975 pKa = 4.66YY976 pKa = 10.72YY977 pKa = 10.89GSGGSGSSGSNTLILGAPGFIAGDD1001 pKa = 3.92IILDD1005 pKa = 3.53GDD1007 pKa = 4.2ANVDD1011 pKa = 3.47VTLTSGAGNSVAWTFDD1027 pKa = 3.5DD1028 pKa = 4.23SGSNGPSLPVMTDD1041 pKa = 3.25GPLPWFTDD1049 pKa = 3.76SNSGGSTWATIDD1061 pKa = 3.28PTAFAAAHH1069 pKa = 5.54NQVADD1074 pKa = 4.04LAGMASGMQFGSMGQDD1090 pKa = 3.28DD1091 pKa = 4.58SGMASGDD1098 pKa = 3.63PMTSMDD1104 pKa = 6.52DD1105 pKa = 3.34GGLWLAIEE1113 pKa = 4.87AGEE1116 pKa = 4.54SDD1118 pKa = 3.69YY1119 pKa = 11.8DD1120 pKa = 4.01GDD1122 pKa = 4.88GVTTLDD1128 pKa = 4.54QEE1130 pKa = 4.57TTLTGIAAGYY1140 pKa = 8.01SAKK1143 pKa = 10.47LNPEE1147 pKa = 3.94TRR1149 pKa = 11.84VGAMLGYY1156 pKa = 9.93GKK1158 pKa = 10.17SEE1160 pKa = 4.62LEE1162 pKa = 3.78VDD1164 pKa = 4.03SNIGIATVKK1173 pKa = 10.33SYY1175 pKa = 10.96EE1176 pKa = 4.14SEE1178 pKa = 3.7QDD1180 pKa = 3.7GIFGGIYY1187 pKa = 10.23GRR1189 pKa = 11.84AKK1191 pKa = 10.43LGSIYY1196 pKa = 10.73LDD1198 pKa = 3.46LALNAGWQSHH1208 pKa = 4.87DD1209 pKa = 3.52QARR1212 pKa = 11.84LVNDD1216 pKa = 3.31NLQNDD1221 pKa = 4.18GTDD1224 pKa = 3.8FANASYY1230 pKa = 11.27DD1231 pKa = 3.9SFWFSPEE1238 pKa = 3.58IGVMLPIMVSDD1249 pKa = 4.23TLTVTPNLRR1258 pKa = 11.84ARR1260 pKa = 11.84YY1261 pKa = 8.93VSQSIDD1267 pKa = 3.05GYY1269 pKa = 7.85TEE1271 pKa = 3.99KK1272 pKa = 10.89DD1273 pKa = 3.59SNSNATVDD1281 pKa = 4.84DD1282 pKa = 3.57IDD1284 pKa = 4.42IGVTEE1289 pKa = 4.69LRR1291 pKa = 11.84GGLDD1295 pKa = 3.17VTHH1298 pKa = 6.55RR1299 pKa = 11.84TSVMALTGTIGYY1311 pKa = 7.4MSRR1314 pKa = 11.84SGSGDD1319 pKa = 3.25DD1320 pKa = 3.75SVNITMILDD1329 pKa = 3.83NNDD1332 pKa = 2.68VGFNYY1337 pKa = 10.38SDD1339 pKa = 2.91VDD1341 pKa = 3.37AAYY1344 pKa = 10.41GGISFAYY1351 pKa = 9.29KK1352 pKa = 9.8MNEE1355 pKa = 3.48ATTLKK1360 pKa = 10.5LNASVTVGDD1369 pKa = 5.45DD1370 pKa = 2.76IDD1372 pKa = 4.02AASASASVNMKK1383 pKa = 9.87FF1384 pKa = 4.46
MM1 pKa = 6.88NHH3 pKa = 6.88KK4 pKa = 10.7ASLNQNKK11 pKa = 9.62SVEE14 pKa = 4.26PNSLQGMRR22 pKa = 11.84RR23 pKa = 11.84TATAIAVASVLSVATLPQATAGFSYY48 pKa = 11.02GSGDD52 pKa = 3.69TIAITGPVTAHH63 pKa = 6.29NNDD66 pKa = 2.9GSDD69 pKa = 3.46LVNVNAYY76 pKa = 9.68FADD79 pKa = 5.4DD80 pKa = 3.21ISGSYY85 pKa = 10.48GVLLRR90 pKa = 11.84GDD92 pKa = 4.08SGASSNTITNNGYY105 pKa = 9.93DD106 pKa = 3.72ADD108 pKa = 4.16VSGSAAAIEE117 pKa = 4.41VGYY120 pKa = 10.46IPGPDD125 pKa = 3.48DD126 pKa = 4.1VRR128 pKa = 11.84AHH130 pKa = 6.54NNTITNSGRR139 pKa = 11.84MEE141 pKa = 4.4GSGGRR146 pKa = 11.84AVPTGRR152 pKa = 11.84TGVAPRR158 pKa = 11.84IVITPPGTYY167 pKa = 9.45LQYY170 pKa = 10.85EE171 pKa = 4.63GTVHH175 pKa = 5.58ITAFAAEE182 pKa = 4.41NNTDD186 pKa = 3.39ADD188 pKa = 3.97AEE190 pKa = 4.22VDD192 pKa = 3.54NNLIEE197 pKa = 4.22NSGTISGDD205 pKa = 2.8GTLAVHH211 pKa = 6.82LGSFAYY217 pKa = 10.59VSGSSAHH224 pKa = 6.86KK225 pKa = 9.48GTAEE229 pKa = 3.84VGNNSVNNSGGIVSDD244 pKa = 3.54STGVQFLSIAGGFSGTHH261 pKa = 6.41AEE263 pKa = 4.03ATVNDD268 pKa = 3.85NVVSNAYY275 pKa = 10.38DD276 pKa = 3.5GDD278 pKa = 4.1INGGMLSGIDD288 pKa = 3.5MTAIAATSSSCCGPQGNADD307 pKa = 3.44ASVEE311 pKa = 4.38GNSNTNDD318 pKa = 3.3GYY320 pKa = 11.53AGDD323 pKa = 4.48SLLSGASMNAIAASGYY339 pKa = 9.48MPFARR344 pKa = 11.84ADD346 pKa = 3.49AEE348 pKa = 4.57VNSNEE353 pKa = 3.65ITNAEE358 pKa = 3.98NANIEE363 pKa = 4.15GKK365 pKa = 10.87YY366 pKa = 10.64NGIALRR372 pKa = 11.84AAGEE376 pKa = 4.29DD377 pKa = 2.8IKK379 pKa = 11.43YY380 pKa = 10.78AGDD383 pKa = 3.46VAVNSNMISNDD394 pKa = 2.81GTVFASEE401 pKa = 4.36DD402 pKa = 3.46QFADD406 pKa = 3.64SAVKK410 pKa = 9.98LASYY414 pKa = 11.08GNLANMFSKK423 pKa = 10.77DD424 pKa = 4.13DD425 pKa = 3.6GHH427 pKa = 6.74GLPFANDD434 pKa = 3.05TDD436 pKa = 4.35VNSNTITNSSTGDD449 pKa = 3.52MVNSAWMVRR458 pKa = 11.84DD459 pKa = 3.39SAVSLRR465 pKa = 11.84ADD467 pKa = 3.07SSTGDD472 pKa = 3.24RR473 pKa = 11.84YY474 pKa = 9.82DD475 pKa = 3.54TPFSGSSSAEE485 pKa = 3.88VNSNSIEE492 pKa = 3.77NHH494 pKa = 4.91GFIGSFAQYY503 pKa = 10.77NFGGGIEE510 pKa = 4.11LTADD514 pKa = 3.38AGGDD518 pKa = 3.36AYY520 pKa = 10.12EE521 pKa = 4.12YY522 pKa = 10.67DD523 pKa = 4.41YY524 pKa = 11.21ISSGGFIDD532 pKa = 3.4HH533 pKa = 6.75EE534 pKa = 4.25EE535 pKa = 4.41RR536 pKa = 11.84IGVSAGTADD545 pKa = 3.6VNKK548 pKa = 8.84NTVSNTSSDD557 pKa = 3.75TIAVGGYY564 pKa = 8.99VLFDD568 pKa = 3.82ADD570 pKa = 5.01GIEE573 pKa = 4.33LNADD577 pKa = 3.29ASASAEE583 pKa = 4.19TYY585 pKa = 10.33IDD587 pKa = 3.2ISGGFSDD594 pKa = 5.07ASGSADD600 pKa = 3.08SLAFAGDD607 pKa = 3.31ADD609 pKa = 4.05VNNNQVEE616 pKa = 4.27NSGSIEE622 pKa = 3.76VTGYY626 pKa = 9.55MGEE629 pKa = 4.15DD630 pKa = 3.14SQGIVLAADD639 pKa = 3.44AWAYY643 pKa = 10.87ANSDD647 pKa = 3.12VTAQSSGSSSADD659 pKa = 3.23SFGMASGGTTFVNNNDD675 pKa = 3.92VINSGDD681 pKa = 3.41ILTSGEE687 pKa = 4.56DD688 pKa = 3.66FPSQYY693 pKa = 9.34MQGILLTSSASAFAYY708 pKa = 10.53SEE710 pKa = 4.04ATAVSGGGNGDD721 pKa = 3.43ASGGATAMAGTAEE734 pKa = 4.03VHH736 pKa = 6.16NNNVVNSGTISTDD749 pKa = 3.33GEE751 pKa = 4.19DD752 pKa = 3.23SSGIEE757 pKa = 3.68LSAFAWADD765 pKa = 3.04ADD767 pKa = 3.72VDD769 pKa = 4.3LSGSSTTDD777 pKa = 2.58SDD779 pKa = 3.61GTTASGGFASVANNTITNSGDD800 pKa = 2.55ITTAGGRR807 pKa = 11.84GASSTGILLYY817 pKa = 10.85ADD819 pKa = 3.66VSAWAEE825 pKa = 4.03RR826 pKa = 11.84FSDD829 pKa = 3.84DD830 pKa = 3.95TDD832 pKa = 4.16GSDD835 pKa = 3.19VARR838 pKa = 11.84AGIAEE843 pKa = 4.06ITGNSVDD850 pKa = 3.44NSGVIDD856 pKa = 3.44AWEE859 pKa = 4.97GIVLEE864 pKa = 4.47ARR866 pKa = 11.84ASAFADD872 pKa = 3.22NYY874 pKa = 10.31ARR876 pKa = 11.84SGEE879 pKa = 4.23AYY881 pKa = 9.24LTDD884 pKa = 3.46NTITNSGTIIADD896 pKa = 3.4NVGIAIRR903 pKa = 11.84GEE905 pKa = 3.92ASGSVINTIFTHH917 pKa = 6.34GNSVDD922 pKa = 3.41NSGSMEE928 pKa = 4.14VYY930 pKa = 9.6NGSGIIVGEE939 pKa = 4.11EE940 pKa = 4.03APSGSGSYY948 pKa = 8.53TWDD951 pKa = 3.08NTINNTGTILAGEE964 pKa = 4.21DD965 pKa = 3.53TAIGIYY971 pKa = 10.53GDD973 pKa = 3.45AEE975 pKa = 4.66YY976 pKa = 10.72YY977 pKa = 10.89GSGGSGSSGSNTLILGAPGFIAGDD1001 pKa = 3.92IILDD1005 pKa = 3.53GDD1007 pKa = 4.2ANVDD1011 pKa = 3.47VTLTSGAGNSVAWTFDD1027 pKa = 3.5DD1028 pKa = 4.23SGSNGPSLPVMTDD1041 pKa = 3.25GPLPWFTDD1049 pKa = 3.76SNSGGSTWATIDD1061 pKa = 3.28PTAFAAAHH1069 pKa = 5.54NQVADD1074 pKa = 4.04LAGMASGMQFGSMGQDD1090 pKa = 3.28DD1091 pKa = 4.58SGMASGDD1098 pKa = 3.63PMTSMDD1104 pKa = 6.52DD1105 pKa = 3.34GGLWLAIEE1113 pKa = 4.87AGEE1116 pKa = 4.54SDD1118 pKa = 3.69YY1119 pKa = 11.8DD1120 pKa = 4.01GDD1122 pKa = 4.88GVTTLDD1128 pKa = 4.54QEE1130 pKa = 4.57TTLTGIAAGYY1140 pKa = 8.01SAKK1143 pKa = 10.47LNPEE1147 pKa = 3.94TRR1149 pKa = 11.84VGAMLGYY1156 pKa = 9.93GKK1158 pKa = 10.17SEE1160 pKa = 4.62LEE1162 pKa = 3.78VDD1164 pKa = 4.03SNIGIATVKK1173 pKa = 10.33SYY1175 pKa = 10.96EE1176 pKa = 4.14SEE1178 pKa = 3.7QDD1180 pKa = 3.7GIFGGIYY1187 pKa = 10.23GRR1189 pKa = 11.84AKK1191 pKa = 10.43LGSIYY1196 pKa = 10.73LDD1198 pKa = 3.46LALNAGWQSHH1208 pKa = 4.87DD1209 pKa = 3.52QARR1212 pKa = 11.84LVNDD1216 pKa = 3.31NLQNDD1221 pKa = 4.18GTDD1224 pKa = 3.8FANASYY1230 pKa = 11.27DD1231 pKa = 3.9SFWFSPEE1238 pKa = 3.58IGVMLPIMVSDD1249 pKa = 4.23TLTVTPNLRR1258 pKa = 11.84ARR1260 pKa = 11.84YY1261 pKa = 8.93VSQSIDD1267 pKa = 3.05GYY1269 pKa = 7.85TEE1271 pKa = 3.99KK1272 pKa = 10.89DD1273 pKa = 3.59SNSNATVDD1281 pKa = 4.84DD1282 pKa = 3.57IDD1284 pKa = 4.42IGVTEE1289 pKa = 4.69LRR1291 pKa = 11.84GGLDD1295 pKa = 3.17VTHH1298 pKa = 6.55RR1299 pKa = 11.84TSVMALTGTIGYY1311 pKa = 7.4MSRR1314 pKa = 11.84SGSGDD1319 pKa = 3.25DD1320 pKa = 3.75SVNITMILDD1329 pKa = 3.83NNDD1332 pKa = 2.68VGFNYY1337 pKa = 10.38SDD1339 pKa = 2.91VDD1341 pKa = 3.37AAYY1344 pKa = 10.41GGISFAYY1351 pKa = 9.29KK1352 pKa = 9.8MNEE1355 pKa = 3.48ATTLKK1360 pKa = 10.5LNASVTVGDD1369 pKa = 5.45DD1370 pKa = 2.76IDD1372 pKa = 4.02AASASASVNMKK1383 pKa = 9.87FF1384 pKa = 4.46
Molecular weight: 140.64 kDa
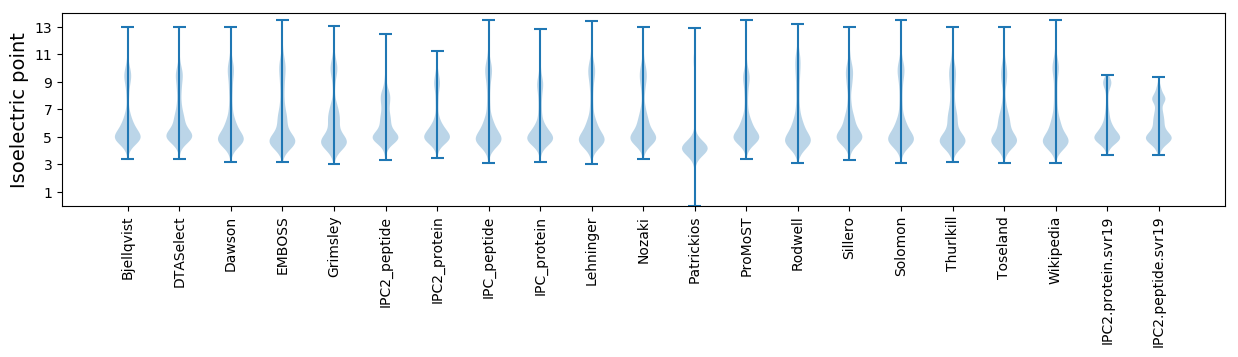
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B0HBU6|A0A0B0HBU6_SOVGS EAL/GGDEF domain-containing protein OS=Solemya velum gill symbiont OX=2340 GN=JV46_11790 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84IKK11 pKa = 10.5RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.95GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.42GGRR28 pKa = 11.84KK29 pKa = 9.17VINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.2GRR39 pKa = 11.84ARR41 pKa = 11.84LAGG44 pKa = 3.63
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84IKK11 pKa = 10.5RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.95GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.42GGRR28 pKa = 11.84KK29 pKa = 9.17VINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.2GRR39 pKa = 11.84ARR41 pKa = 11.84LAGG44 pKa = 3.63
Molecular weight: 5.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
796070 |
19 |
2861 |
299.6 |
33.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.036 ± 0.052 | 1.018 ± 0.02 |
5.898 ± 0.042 | 6.902 ± 0.054 |
3.767 ± 0.034 | 7.485 ± 0.038 |
2.377 ± 0.026 | 6.168 ± 0.034 |
4.515 ± 0.045 | 10.287 ± 0.065 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.827 ± 0.023 | 3.496 ± 0.031 |
4.328 ± 0.029 | 3.82 ± 0.031 |
5.614 ± 0.036 | 6.293 ± 0.043 |
5.144 ± 0.026 | 6.986 ± 0.043 |
1.276 ± 0.024 | 2.765 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
