
Geodermatophilus dictyosporus
Taxonomy:
Average proteome isoelectric point is 6.22
Get precalculated fractions of proteins
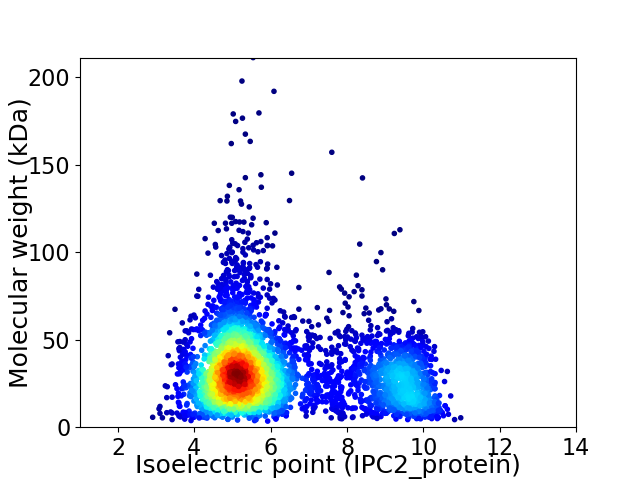
Virtual 2D-PAGE plot for 4366 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I5LXP0|A0A1I5LXP0_9ACTN Carbon-monoxide dehydrogenase large subunit OS=Geodermatophilus dictyosporus OX=1523247 GN=SAMN05660464_1960 PE=4 SV=1
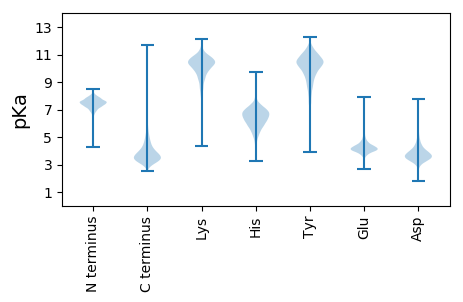
MM1 pKa = 7.85RR2 pKa = 11.84VRR4 pKa = 11.84PPAVAALLALALAATGCSTVVAGVASPPPGGLRR37 pKa = 11.84TDD39 pKa = 3.29ATDD42 pKa = 3.49ATDD45 pKa = 4.01ADD47 pKa = 4.46LTVHH51 pKa = 6.6LAEE54 pKa = 5.92AGDD57 pKa = 3.75EE58 pKa = 4.17TDD60 pKa = 4.32RR61 pKa = 11.84IARR64 pKa = 11.84NALADD69 pKa = 3.56VLTYY73 pKa = 9.93WDD75 pKa = 3.39RR76 pKa = 11.84TYY78 pKa = 11.3PEE80 pKa = 4.12VFGGGFGPVDD90 pKa = 3.3GGFWSIDD97 pKa = 3.4PDD99 pKa = 3.7EE100 pKa = 4.99TDD102 pKa = 4.52PADD105 pKa = 5.18LPDD108 pKa = 4.85GPCFPDD114 pKa = 4.55DD115 pKa = 4.91VEE117 pKa = 5.55DD118 pKa = 4.23LADD121 pKa = 3.52NAYY124 pKa = 10.12YY125 pKa = 10.74CPDD128 pKa = 3.84EE129 pKa = 5.02DD130 pKa = 4.41LVVYY134 pKa = 10.46DD135 pKa = 4.31RR136 pKa = 11.84AWMAEE141 pKa = 3.84LAGDD145 pKa = 3.67YY146 pKa = 11.06GPFVVAEE153 pKa = 4.34IMAHH157 pKa = 6.32EE158 pKa = 4.6IAHH161 pKa = 6.66AVQAHH166 pKa = 6.73AGLDD170 pKa = 3.66DD171 pKa = 4.22PSIVAEE177 pKa = 4.24TQAEE181 pKa = 4.46CFAGAWTRR189 pKa = 11.84WVVRR193 pKa = 11.84GNAAHH198 pKa = 6.55FSVRR202 pKa = 11.84PKK204 pKa = 10.82EE205 pKa = 3.93LDD207 pKa = 3.63PYY209 pKa = 11.04LLGYY213 pKa = 10.47LYY215 pKa = 10.83FGDD218 pKa = 4.37EE219 pKa = 4.51VGSSPDD225 pKa = 3.38AEE227 pKa = 4.42DD228 pKa = 3.14AHH230 pKa = 7.9GSVFDD235 pKa = 3.75QLSAFQEE242 pKa = 5.47GYY244 pKa = 11.07ADD246 pKa = 4.26GPVACAAFDD255 pKa = 3.54SSRR258 pKa = 11.84LFTEE262 pKa = 4.41EE263 pKa = 4.67EE264 pKa = 3.87FDD266 pKa = 3.74GDD268 pKa = 3.64EE269 pKa = 4.25AATGGDD275 pKa = 3.57LSYY278 pKa = 10.6EE279 pKa = 4.01AVVAGTGDD287 pKa = 4.23LLTAFWDD294 pKa = 3.61QALTRR299 pKa = 11.84GFPGTDD305 pKa = 3.21PLGGLLDD312 pKa = 4.46APDD315 pKa = 4.04LRR317 pKa = 11.84AADD320 pKa = 3.86GTGTVCGGGGEE331 pKa = 5.62LDD333 pKa = 3.74LQYY336 pKa = 11.22CPVDD340 pKa = 3.48DD341 pKa = 3.87SVRR344 pKa = 11.84YY345 pKa = 10.19DD346 pKa = 3.39GADD349 pKa = 3.43LLEE352 pKa = 4.17PAYY355 pKa = 10.74AEE357 pKa = 4.19VGDD360 pKa = 4.14FAVPTLLGLPYY371 pKa = 11.09AMAVRR376 pKa = 11.84EE377 pKa = 3.98QRR379 pKa = 11.84GLSVDD384 pKa = 3.53DD385 pKa = 4.12AAAVTGSVCATGWLVRR401 pKa = 11.84EE402 pKa = 4.11VHH404 pKa = 6.33RR405 pKa = 11.84GSVDD409 pKa = 3.42GLDD412 pKa = 3.9LALSPGDD419 pKa = 3.39VDD421 pKa = 3.6EE422 pKa = 4.97AAVVLLRR429 pKa = 11.84YY430 pKa = 8.44ATEE433 pKa = 4.04EE434 pKa = 4.09TVVPVPGLSGFEE446 pKa = 4.02LVDD449 pKa = 3.36DD450 pKa = 4.74FRR452 pKa = 11.84RR453 pKa = 11.84GFVDD457 pKa = 4.65GGAACGFF464 pKa = 4.12
MM1 pKa = 7.85RR2 pKa = 11.84VRR4 pKa = 11.84PPAVAALLALALAATGCSTVVAGVASPPPGGLRR37 pKa = 11.84TDD39 pKa = 3.29ATDD42 pKa = 3.49ATDD45 pKa = 4.01ADD47 pKa = 4.46LTVHH51 pKa = 6.6LAEE54 pKa = 5.92AGDD57 pKa = 3.75EE58 pKa = 4.17TDD60 pKa = 4.32RR61 pKa = 11.84IARR64 pKa = 11.84NALADD69 pKa = 3.56VLTYY73 pKa = 9.93WDD75 pKa = 3.39RR76 pKa = 11.84TYY78 pKa = 11.3PEE80 pKa = 4.12VFGGGFGPVDD90 pKa = 3.3GGFWSIDD97 pKa = 3.4PDD99 pKa = 3.7EE100 pKa = 4.99TDD102 pKa = 4.52PADD105 pKa = 5.18LPDD108 pKa = 4.85GPCFPDD114 pKa = 4.55DD115 pKa = 4.91VEE117 pKa = 5.55DD118 pKa = 4.23LADD121 pKa = 3.52NAYY124 pKa = 10.12YY125 pKa = 10.74CPDD128 pKa = 3.84EE129 pKa = 5.02DD130 pKa = 4.41LVVYY134 pKa = 10.46DD135 pKa = 4.31RR136 pKa = 11.84AWMAEE141 pKa = 3.84LAGDD145 pKa = 3.67YY146 pKa = 11.06GPFVVAEE153 pKa = 4.34IMAHH157 pKa = 6.32EE158 pKa = 4.6IAHH161 pKa = 6.66AVQAHH166 pKa = 6.73AGLDD170 pKa = 3.66DD171 pKa = 4.22PSIVAEE177 pKa = 4.24TQAEE181 pKa = 4.46CFAGAWTRR189 pKa = 11.84WVVRR193 pKa = 11.84GNAAHH198 pKa = 6.55FSVRR202 pKa = 11.84PKK204 pKa = 10.82EE205 pKa = 3.93LDD207 pKa = 3.63PYY209 pKa = 11.04LLGYY213 pKa = 10.47LYY215 pKa = 10.83FGDD218 pKa = 4.37EE219 pKa = 4.51VGSSPDD225 pKa = 3.38AEE227 pKa = 4.42DD228 pKa = 3.14AHH230 pKa = 7.9GSVFDD235 pKa = 3.75QLSAFQEE242 pKa = 5.47GYY244 pKa = 11.07ADD246 pKa = 4.26GPVACAAFDD255 pKa = 3.54SSRR258 pKa = 11.84LFTEE262 pKa = 4.41EE263 pKa = 4.67EE264 pKa = 3.87FDD266 pKa = 3.74GDD268 pKa = 3.64EE269 pKa = 4.25AATGGDD275 pKa = 3.57LSYY278 pKa = 10.6EE279 pKa = 4.01AVVAGTGDD287 pKa = 4.23LLTAFWDD294 pKa = 3.61QALTRR299 pKa = 11.84GFPGTDD305 pKa = 3.21PLGGLLDD312 pKa = 4.46APDD315 pKa = 4.04LRR317 pKa = 11.84AADD320 pKa = 3.86GTGTVCGGGGEE331 pKa = 5.62LDD333 pKa = 3.74LQYY336 pKa = 11.22CPVDD340 pKa = 3.48DD341 pKa = 3.87SVRR344 pKa = 11.84YY345 pKa = 10.19DD346 pKa = 3.39GADD349 pKa = 3.43LLEE352 pKa = 4.17PAYY355 pKa = 10.74AEE357 pKa = 4.19VGDD360 pKa = 4.14FAVPTLLGLPYY371 pKa = 11.09AMAVRR376 pKa = 11.84EE377 pKa = 3.98QRR379 pKa = 11.84GLSVDD384 pKa = 3.53DD385 pKa = 4.12AAAVTGSVCATGWLVRR401 pKa = 11.84EE402 pKa = 4.11VHH404 pKa = 6.33RR405 pKa = 11.84GSVDD409 pKa = 3.42GLDD412 pKa = 3.9LALSPGDD419 pKa = 3.39VDD421 pKa = 3.6EE422 pKa = 4.97AAVVLLRR429 pKa = 11.84YY430 pKa = 8.44ATEE433 pKa = 4.04EE434 pKa = 4.09TVVPVPGLSGFEE446 pKa = 4.02LVDD449 pKa = 3.36DD450 pKa = 4.74FRR452 pKa = 11.84RR453 pKa = 11.84GFVDD457 pKa = 4.65GGAACGFF464 pKa = 4.12
Molecular weight: 48.61 kDa
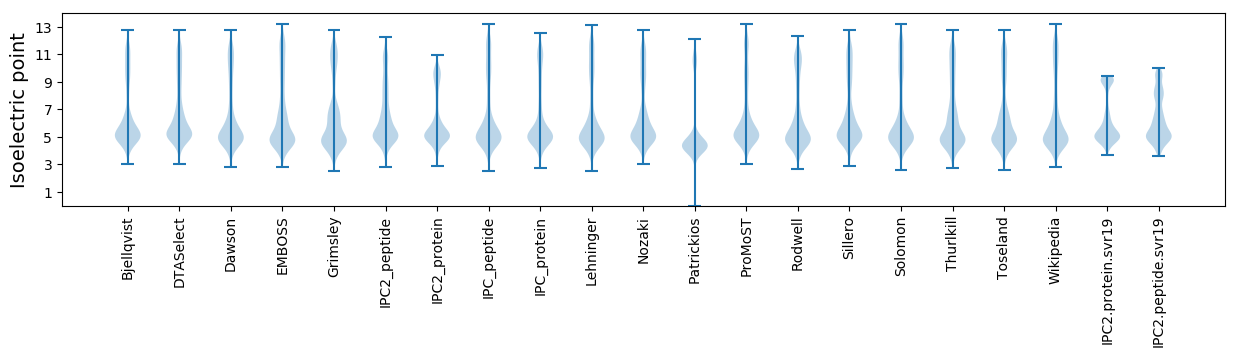
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I5S9Y9|A0A1I5S9Y9_9ACTN DNA gyrase subunit B OS=Geodermatophilus dictyosporus OX=1523247 GN=gyrB PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84SKK15 pKa = 8.58THH17 pKa = 5.46GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILAGRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 10.08GRR40 pKa = 11.84EE41 pKa = 3.61KK42 pKa = 10.93LSAA45 pKa = 3.78
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84SKK15 pKa = 8.58THH17 pKa = 5.46GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILAGRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 10.08GRR40 pKa = 11.84EE41 pKa = 3.61KK42 pKa = 10.93LSAA45 pKa = 3.78
Molecular weight: 5.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1400948 |
32 |
2011 |
320.9 |
33.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.832 ± 0.071 | 0.668 ± 0.01 |
6.255 ± 0.027 | 5.479 ± 0.035 |
2.439 ± 0.021 | 9.917 ± 0.032 |
1.989 ± 0.017 | 2.231 ± 0.028 |
1.138 ± 0.021 | 10.811 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.43 ± 0.014 | 1.264 ± 0.015 |
6.494 ± 0.034 | 2.558 ± 0.018 |
8.515 ± 0.043 | 4.55 ± 0.025 |
5.984 ± 0.031 | 10.283 ± 0.034 |
1.48 ± 0.018 | 1.683 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
