
Streptomyces sp. AgN23
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; unclassified Streptomyces
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
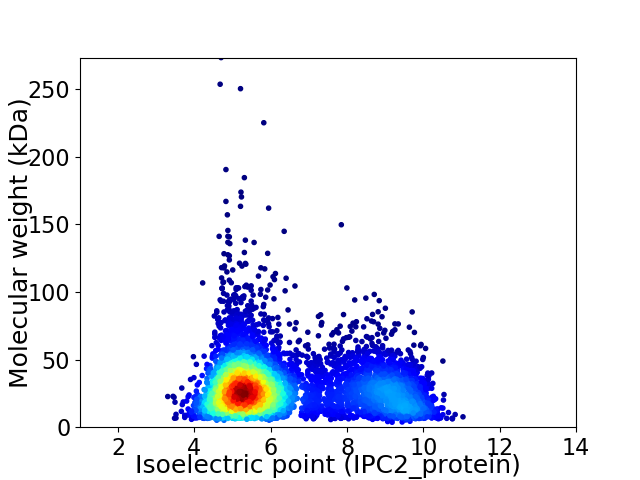
Virtual 2D-PAGE plot for 5582 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6F7VQ29|A0A6F7VQ29_9ACTN NADH-quinone oxidoreductase subunit A OS=Streptomyces sp. AgN23 OX=1188315 GN=nuoA PE=3 SV=1
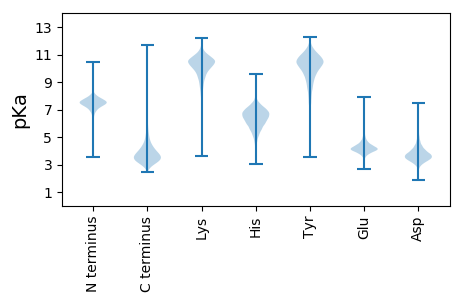
MM1 pKa = 7.89RR2 pKa = 11.84MYY4 pKa = 10.32RR5 pKa = 11.84TMAAAATLALALGGAALAAPAAQAAATTSGSLVHH39 pKa = 7.09EE40 pKa = 5.54DD41 pKa = 3.54GEE43 pKa = 4.49LWYY46 pKa = 10.53KK47 pKa = 10.45AAAGQKK53 pKa = 9.77NNLSVSEE60 pKa = 4.2QIVSRR65 pKa = 11.84GEE67 pKa = 3.71FEE69 pKa = 4.1EE70 pKa = 5.66YY71 pKa = 10.54YY72 pKa = 11.25VLTFRR77 pKa = 11.84DD78 pKa = 3.77NGDD81 pKa = 2.91ITIDD85 pKa = 3.91AEE87 pKa = 3.98AADD90 pKa = 4.52WDD92 pKa = 4.01EE93 pKa = 4.28CTYY96 pKa = 8.96PTAGDD101 pKa = 3.56HH102 pKa = 6.36TVAQCAVLIPQNSDD116 pKa = 3.21DD117 pKa = 3.95SDD119 pKa = 4.15NYY121 pKa = 11.38DD122 pKa = 3.0VDD124 pKa = 5.67LGDD127 pKa = 4.68GDD129 pKa = 4.54DD130 pKa = 4.37TIKK133 pKa = 11.07LDD135 pKa = 4.25ADD137 pKa = 3.43SSAYY141 pKa = 10.39AGIHH145 pKa = 5.97GGAGDD150 pKa = 5.29DD151 pKa = 3.73VLKK154 pKa = 11.28GNAAPMFYY162 pKa = 10.96GEE164 pKa = 5.66DD165 pKa = 3.57GDD167 pKa = 4.6DD168 pKa = 3.87QIDD171 pKa = 3.77GGGGVMGFGAYY182 pKa = 10.05GGDD185 pKa = 3.69GDD187 pKa = 5.06DD188 pKa = 4.57TITNCVQEE196 pKa = 4.3CHH198 pKa = 6.56GDD200 pKa = 3.57AGDD203 pKa = 3.75DD204 pKa = 4.08TITGGAEE211 pKa = 3.76DD212 pKa = 4.17NILRR216 pKa = 11.84GGTGKK221 pKa = 10.54DD222 pKa = 2.76ILRR225 pKa = 11.84GGKK228 pKa = 9.68GADD231 pKa = 4.08AIYY234 pKa = 10.67GDD236 pKa = 4.03QDD238 pKa = 4.34DD239 pKa = 4.09DD240 pKa = 3.86TLYY243 pKa = 11.47GEE245 pKa = 5.37DD246 pKa = 4.59GNDD249 pKa = 3.43TIYY252 pKa = 11.26GNSGNDD258 pKa = 3.47VLWGGRR264 pKa = 11.84GTDD267 pKa = 3.64TLSGGPGRR275 pKa = 11.84NEE277 pKa = 3.68VHH279 pKa = 6.65QDD281 pKa = 2.84
MM1 pKa = 7.89RR2 pKa = 11.84MYY4 pKa = 10.32RR5 pKa = 11.84TMAAAATLALALGGAALAAPAAQAAATTSGSLVHH39 pKa = 7.09EE40 pKa = 5.54DD41 pKa = 3.54GEE43 pKa = 4.49LWYY46 pKa = 10.53KK47 pKa = 10.45AAAGQKK53 pKa = 9.77NNLSVSEE60 pKa = 4.2QIVSRR65 pKa = 11.84GEE67 pKa = 3.71FEE69 pKa = 4.1EE70 pKa = 5.66YY71 pKa = 10.54YY72 pKa = 11.25VLTFRR77 pKa = 11.84DD78 pKa = 3.77NGDD81 pKa = 2.91ITIDD85 pKa = 3.91AEE87 pKa = 3.98AADD90 pKa = 4.52WDD92 pKa = 4.01EE93 pKa = 4.28CTYY96 pKa = 8.96PTAGDD101 pKa = 3.56HH102 pKa = 6.36TVAQCAVLIPQNSDD116 pKa = 3.21DD117 pKa = 3.95SDD119 pKa = 4.15NYY121 pKa = 11.38DD122 pKa = 3.0VDD124 pKa = 5.67LGDD127 pKa = 4.68GDD129 pKa = 4.54DD130 pKa = 4.37TIKK133 pKa = 11.07LDD135 pKa = 4.25ADD137 pKa = 3.43SSAYY141 pKa = 10.39AGIHH145 pKa = 5.97GGAGDD150 pKa = 5.29DD151 pKa = 3.73VLKK154 pKa = 11.28GNAAPMFYY162 pKa = 10.96GEE164 pKa = 5.66DD165 pKa = 3.57GDD167 pKa = 4.6DD168 pKa = 3.87QIDD171 pKa = 3.77GGGGVMGFGAYY182 pKa = 10.05GGDD185 pKa = 3.69GDD187 pKa = 5.06DD188 pKa = 4.57TITNCVQEE196 pKa = 4.3CHH198 pKa = 6.56GDD200 pKa = 3.57AGDD203 pKa = 3.75DD204 pKa = 4.08TITGGAEE211 pKa = 3.76DD212 pKa = 4.17NILRR216 pKa = 11.84GGTGKK221 pKa = 10.54DD222 pKa = 2.76ILRR225 pKa = 11.84GGKK228 pKa = 9.68GADD231 pKa = 4.08AIYY234 pKa = 10.67GDD236 pKa = 4.03QDD238 pKa = 4.34DD239 pKa = 4.09DD240 pKa = 3.86TLYY243 pKa = 11.47GEE245 pKa = 5.37DD246 pKa = 4.59GNDD249 pKa = 3.43TIYY252 pKa = 11.26GNSGNDD258 pKa = 3.47VLWGGRR264 pKa = 11.84GTDD267 pKa = 3.64TLSGGPGRR275 pKa = 11.84NEE277 pKa = 3.68VHH279 pKa = 6.65QDD281 pKa = 2.84
Molecular weight: 28.93 kDa
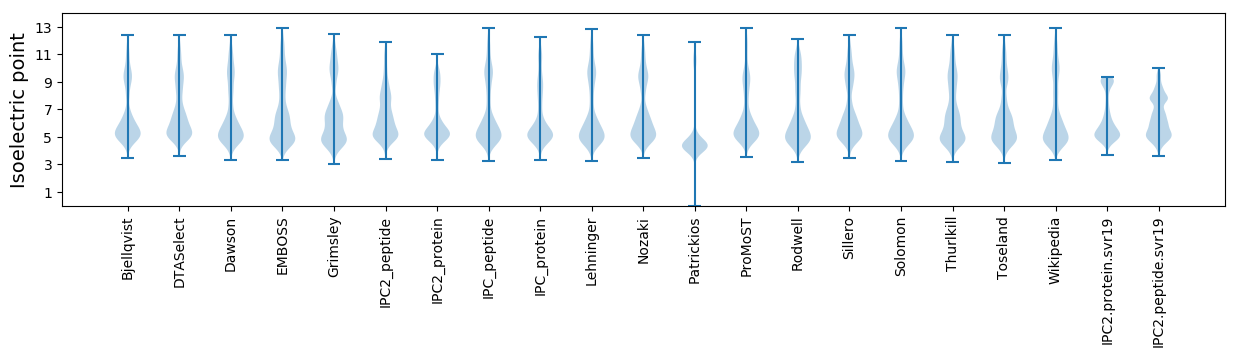
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6F7W0M9|A0A6F7W0M9_9ACTN ABC transporter OS=Streptomyces sp. AgN23 OX=1188315 GN=AS97_37335 PE=4 SV=1
MM1 pKa = 7.38SPRR4 pKa = 11.84RR5 pKa = 11.84NRR7 pKa = 11.84PRR9 pKa = 11.84GGEE12 pKa = 3.88KK13 pKa = 10.23PVGHH17 pKa = 6.72EE18 pKa = 3.71SANRR22 pKa = 11.84YY23 pKa = 9.37GLDD26 pKa = 5.08RR27 pKa = 11.84MEE29 pKa = 4.01TWQGEE34 pKa = 4.39EE35 pKa = 3.8WVVRR39 pKa = 11.84QVGGGAAAKK48 pKa = 9.9HH49 pKa = 5.06YY50 pKa = 9.18RR51 pKa = 11.84CPGCDD56 pKa = 3.24QEE58 pKa = 4.75IPPGVAHH65 pKa = 5.96VVAWQQHH72 pKa = 4.58MGADD76 pKa = 5.12DD77 pKa = 3.31RR78 pKa = 11.84RR79 pKa = 11.84HH80 pKa = 3.84WHH82 pKa = 5.64KK83 pKa = 11.08ACWSARR89 pKa = 11.84DD90 pKa = 3.54RR91 pKa = 11.84RR92 pKa = 11.84SAKK95 pKa = 9.7LQRR98 pKa = 11.84SRR100 pKa = 11.84NAPRR104 pKa = 11.84YY105 pKa = 8.88
MM1 pKa = 7.38SPRR4 pKa = 11.84RR5 pKa = 11.84NRR7 pKa = 11.84PRR9 pKa = 11.84GGEE12 pKa = 3.88KK13 pKa = 10.23PVGHH17 pKa = 6.72EE18 pKa = 3.71SANRR22 pKa = 11.84YY23 pKa = 9.37GLDD26 pKa = 5.08RR27 pKa = 11.84MEE29 pKa = 4.01TWQGEE34 pKa = 4.39EE35 pKa = 3.8WVVRR39 pKa = 11.84QVGGGAAAKK48 pKa = 9.9HH49 pKa = 5.06YY50 pKa = 9.18RR51 pKa = 11.84CPGCDD56 pKa = 3.24QEE58 pKa = 4.75IPPGVAHH65 pKa = 5.96VVAWQQHH72 pKa = 4.58MGADD76 pKa = 5.12DD77 pKa = 3.31RR78 pKa = 11.84RR79 pKa = 11.84HH80 pKa = 3.84WHH82 pKa = 5.64KK83 pKa = 11.08ACWSARR89 pKa = 11.84DD90 pKa = 3.54RR91 pKa = 11.84RR92 pKa = 11.84SAKK95 pKa = 9.7LQRR98 pKa = 11.84SRR100 pKa = 11.84NAPRR104 pKa = 11.84YY105 pKa = 8.88
Molecular weight: 12.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1629245 |
33 |
2605 |
291.9 |
31.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.027 ± 0.043 | 0.796 ± 0.009 |
6.008 ± 0.027 | 5.903 ± 0.037 |
2.834 ± 0.018 | 9.459 ± 0.034 |
2.348 ± 0.017 | 3.487 ± 0.02 |
2.319 ± 0.029 | 10.231 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.897 ± 0.012 | 1.803 ± 0.017 |
5.95 ± 0.026 | 2.763 ± 0.018 |
7.996 ± 0.036 | 5.174 ± 0.027 |
6.053 ± 0.029 | 8.312 ± 0.032 |
1.496 ± 0.013 | 2.144 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
