
Aureimonas sp. Leaf324
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Aurantimonadaceae; Aureimonas; unclassified Aureimonas
Average proteome isoelectric point is 6.38
Get precalculated fractions of proteins
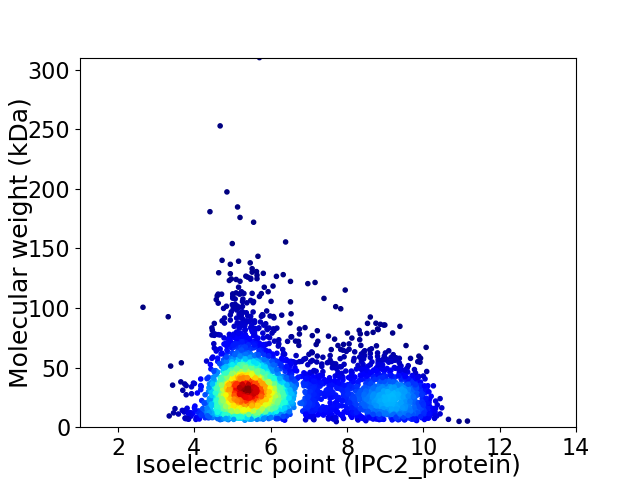
Virtual 2D-PAGE plot for 3989 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q5GG35|A0A0Q5GG35_9RHIZ Formate dehydrogenase OS=Aureimonas sp. Leaf324 OX=1736336 GN=ASF65_10815 PE=3 SV=1
MM1 pKa = 7.68AATPLTGSKK10 pKa = 9.52WGDD13 pKa = 3.24SGVAGTPAGAVTWSFDD29 pKa = 3.78TTGSRR34 pKa = 11.84YY35 pKa = 9.27YY36 pKa = 10.84AYY38 pKa = 10.41DD39 pKa = 4.58DD40 pKa = 4.66IIASEE45 pKa = 5.29GYY47 pKa = 9.9RR48 pKa = 11.84SLVRR52 pKa = 11.84QAFDD56 pKa = 2.6AWEE59 pKa = 4.07KK60 pKa = 10.14VAAIDD65 pKa = 3.87FVEE68 pKa = 4.67VGSGPAEE75 pKa = 3.85IQVGLDD81 pKa = 3.63AIDD84 pKa = 3.88GRR86 pKa = 11.84GGTIGQAVWYY96 pKa = 9.9SRR98 pKa = 11.84GSVTTHH104 pKa = 6.65AEE106 pKa = 3.74IAVDD110 pKa = 4.66SAEE113 pKa = 4.12TWSPFVPNGSNFFGMMVHH131 pKa = 7.2EE132 pKa = 5.08IGHH135 pKa = 6.79AIGLEE140 pKa = 4.08HH141 pKa = 7.31SDD143 pKa = 4.02DD144 pKa = 4.54PSSIMYY150 pKa = 8.6PIASAYY156 pKa = 8.76MAFSNEE162 pKa = 3.75DD163 pKa = 3.1LAAIQALYY171 pKa = 10.62GPAAVAGRR179 pKa = 11.84LLYY182 pKa = 8.99GTSAPDD188 pKa = 3.71LLVGASGNDD197 pKa = 3.13TVYY200 pKa = 10.94GFEE203 pKa = 4.47GADD206 pKa = 3.36NLFGYY211 pKa = 10.26AGNDD215 pKa = 4.03LIAGNQGNDD224 pKa = 3.43QIFGGQGNDD233 pKa = 2.56IVYY236 pKa = 10.49GGRR239 pKa = 11.84DD240 pKa = 3.21QDD242 pKa = 4.02TVSGNLGNDD251 pKa = 3.17TVYY254 pKa = 11.5GDD256 pKa = 4.37LGNDD260 pKa = 3.3TVYY263 pKa = 10.99GGQGDD268 pKa = 4.07DD269 pKa = 3.34LVYY272 pKa = 10.78GGRR275 pKa = 11.84GDD277 pKa = 4.72DD278 pKa = 4.57LVSGDD283 pKa = 4.74LGNDD287 pKa = 3.46TLVGGFGNDD296 pKa = 2.92WLAGRR301 pKa = 11.84AGADD305 pKa = 3.06VFVFEE310 pKa = 5.47AGQGDD315 pKa = 4.44DD316 pKa = 4.63VIVDD320 pKa = 4.08FNAAEE325 pKa = 4.48GDD327 pKa = 3.74RR328 pKa = 11.84LVLSGQTYY336 pKa = 6.85TTAEE340 pKa = 4.2VNGSEE345 pKa = 4.21VLTLSGGGTITLLGVGTAPLEE366 pKa = 4.33AAALAA371 pKa = 4.32
MM1 pKa = 7.68AATPLTGSKK10 pKa = 9.52WGDD13 pKa = 3.24SGVAGTPAGAVTWSFDD29 pKa = 3.78TTGSRR34 pKa = 11.84YY35 pKa = 9.27YY36 pKa = 10.84AYY38 pKa = 10.41DD39 pKa = 4.58DD40 pKa = 4.66IIASEE45 pKa = 5.29GYY47 pKa = 9.9RR48 pKa = 11.84SLVRR52 pKa = 11.84QAFDD56 pKa = 2.6AWEE59 pKa = 4.07KK60 pKa = 10.14VAAIDD65 pKa = 3.87FVEE68 pKa = 4.67VGSGPAEE75 pKa = 3.85IQVGLDD81 pKa = 3.63AIDD84 pKa = 3.88GRR86 pKa = 11.84GGTIGQAVWYY96 pKa = 9.9SRR98 pKa = 11.84GSVTTHH104 pKa = 6.65AEE106 pKa = 3.74IAVDD110 pKa = 4.66SAEE113 pKa = 4.12TWSPFVPNGSNFFGMMVHH131 pKa = 7.2EE132 pKa = 5.08IGHH135 pKa = 6.79AIGLEE140 pKa = 4.08HH141 pKa = 7.31SDD143 pKa = 4.02DD144 pKa = 4.54PSSIMYY150 pKa = 8.6PIASAYY156 pKa = 8.76MAFSNEE162 pKa = 3.75DD163 pKa = 3.1LAAIQALYY171 pKa = 10.62GPAAVAGRR179 pKa = 11.84LLYY182 pKa = 8.99GTSAPDD188 pKa = 3.71LLVGASGNDD197 pKa = 3.13TVYY200 pKa = 10.94GFEE203 pKa = 4.47GADD206 pKa = 3.36NLFGYY211 pKa = 10.26AGNDD215 pKa = 4.03LIAGNQGNDD224 pKa = 3.43QIFGGQGNDD233 pKa = 2.56IVYY236 pKa = 10.49GGRR239 pKa = 11.84DD240 pKa = 3.21QDD242 pKa = 4.02TVSGNLGNDD251 pKa = 3.17TVYY254 pKa = 11.5GDD256 pKa = 4.37LGNDD260 pKa = 3.3TVYY263 pKa = 10.99GGQGDD268 pKa = 4.07DD269 pKa = 3.34LVYY272 pKa = 10.78GGRR275 pKa = 11.84GDD277 pKa = 4.72DD278 pKa = 4.57LVSGDD283 pKa = 4.74LGNDD287 pKa = 3.46TLVGGFGNDD296 pKa = 2.92WLAGRR301 pKa = 11.84AGADD305 pKa = 3.06VFVFEE310 pKa = 5.47AGQGDD315 pKa = 4.44DD316 pKa = 4.63VIVDD320 pKa = 4.08FNAAEE325 pKa = 4.48GDD327 pKa = 3.74RR328 pKa = 11.84LVLSGQTYY336 pKa = 6.85TTAEE340 pKa = 4.2VNGSEE345 pKa = 4.21VLTLSGGGTITLLGVGTAPLEE366 pKa = 4.33AAALAA371 pKa = 4.32
Molecular weight: 38.03 kDa
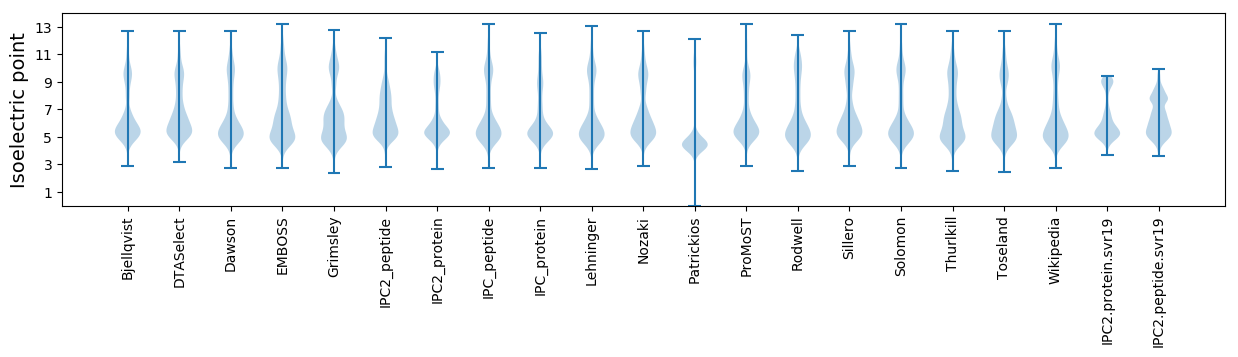
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q5GFP8|A0A0Q5GFP8_9RHIZ Succinyl-diaminopimelate desuccinylase OS=Aureimonas sp. Leaf324 OX=1736336 GN=dapE PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATTGGRR28 pKa = 11.84KK29 pKa = 9.29VIAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATTGGRR28 pKa = 11.84KK29 pKa = 9.29VIAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1266772 |
41 |
2829 |
317.6 |
34.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.32 ± 0.053 | 0.731 ± 0.011 |
5.863 ± 0.033 | 5.919 ± 0.039 |
3.7 ± 0.023 | 8.991 ± 0.041 |
1.893 ± 0.019 | 4.796 ± 0.028 |
2.644 ± 0.035 | 10.213 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.329 ± 0.017 | 2.244 ± 0.021 |
5.245 ± 0.031 | 2.743 ± 0.022 |
7.86 ± 0.039 | 5.308 ± 0.026 |
5.301 ± 0.024 | 7.745 ± 0.034 |
1.191 ± 0.014 | 1.965 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
