
Pseudoalteromonas denitrificans DSM 6059
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Pseudoalteromonadaceae; Pseudoalteromonas; Pseudoalteromonas denitrificans
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
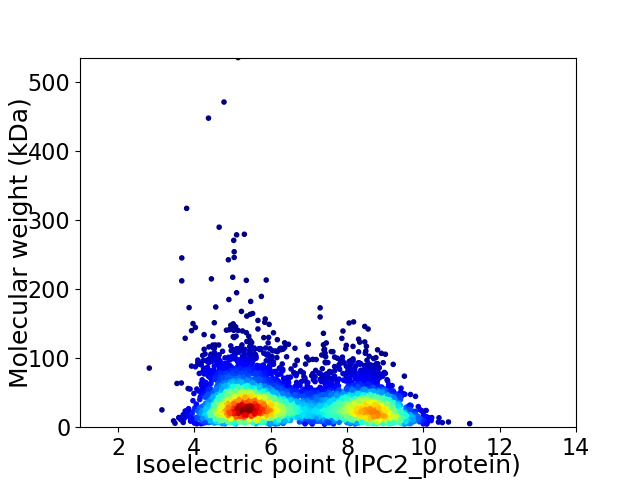
Virtual 2D-PAGE plot for 5332 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I1T2I5|A0A1I1T2I5_9GAMM Biotin-lipoyl like OS=Pseudoalteromonas denitrificans DSM 6059 OX=1123010 GN=SAMN02745724_04771 PE=4 SV=1
MM1 pKa = 7.59SRR3 pKa = 11.84FSPVNFSKK11 pKa = 11.09YY12 pKa = 10.58LLLILTTSAVLFSTPSIAITISDD35 pKa = 3.51VSTTNFSNSDD45 pKa = 3.3HH46 pKa = 6.31YY47 pKa = 11.5QDD49 pKa = 5.55DD50 pKa = 4.24GNSLYY55 pKa = 10.6RR56 pKa = 11.84EE57 pKa = 4.06YY58 pKa = 11.24SSNLDD63 pKa = 2.86ITTVTNNNNDD73 pKa = 4.01AIINGHH79 pKa = 6.19LAWFHH84 pKa = 5.6GMRR87 pKa = 11.84VDD89 pKa = 4.36QPEE92 pKa = 4.54GPNFALIYY100 pKa = 9.0RR101 pKa = 11.84HH102 pKa = 6.13NIGYY106 pKa = 9.18EE107 pKa = 3.48ITFTVNDD114 pKa = 5.12PIEE117 pKa = 4.3EE118 pKa = 4.73GYY120 pKa = 10.46EE121 pKa = 3.73ISIEE125 pKa = 3.91QHH127 pKa = 4.91MKK129 pKa = 10.91GLITVSRR136 pKa = 11.84EE137 pKa = 3.53EE138 pKa = 4.73AISVSATAGLMLGRR152 pKa = 11.84VDD154 pKa = 3.57EE155 pKa = 4.62HH156 pKa = 8.14QGDD159 pKa = 4.53GPIHH163 pKa = 6.36LAGFSISGGGLSVDD177 pKa = 4.12SEE179 pKa = 4.49ATDD182 pKa = 3.65TVQTITIDD190 pKa = 3.49EE191 pKa = 4.59TDD193 pKa = 3.65TQNLATQYY201 pKa = 10.58IGTHH205 pKa = 5.24TFTMSFSSFPSPALVSIFQNSGGGEE230 pKa = 3.84TSIQFGVTSNQDD242 pKa = 2.59AFLYY246 pKa = 11.01GNATQNGLIDD256 pKa = 4.48LTPLGHH262 pKa = 6.9FSQINITSLNLTAPDD277 pKa = 3.68TDD279 pKa = 3.6NDD281 pKa = 4.01GVADD285 pKa = 4.6NIDD288 pKa = 3.65NCVLTSNPDD297 pKa = 3.41QLDD300 pKa = 3.49TDD302 pKa = 4.08GDD304 pKa = 4.5GVGDD308 pKa = 3.69VCDD311 pKa = 3.66NCMEE315 pKa = 4.54TPNSDD320 pKa = 4.37QLDD323 pKa = 3.74TDD325 pKa = 3.49EE326 pKa = 6.42DD327 pKa = 5.07GIGDD331 pKa = 3.77VCDD334 pKa = 3.7NCLDD338 pKa = 3.86TSNPDD343 pKa = 3.28QLDD346 pKa = 3.2IDD348 pKa = 4.0EE349 pKa = 5.64DD350 pKa = 4.94GIGDD354 pKa = 3.77VCDD357 pKa = 3.47NCMDD361 pKa = 4.23TSNPDD366 pKa = 3.2QLDD369 pKa = 3.2IDD371 pKa = 4.0EE372 pKa = 5.64DD373 pKa = 4.94GIGDD377 pKa = 3.77VCDD380 pKa = 3.48NCMDD384 pKa = 4.28TPNSDD389 pKa = 3.56QLDD392 pKa = 3.5IDD394 pKa = 3.78EE395 pKa = 5.67DD396 pKa = 4.94GIGDD400 pKa = 3.74VCDD403 pKa = 3.48NCMGTPNSDD412 pKa = 3.91QLDD415 pKa = 3.74TDD417 pKa = 3.6EE418 pKa = 6.34DD419 pKa = 4.72GVGDD423 pKa = 3.82VCDD426 pKa = 3.83NCVDD430 pKa = 3.8TSNPDD435 pKa = 3.54QVDD438 pKa = 3.43TDD440 pKa = 4.14DD441 pKa = 6.01DD442 pKa = 5.44GIGDD446 pKa = 3.75MCDD449 pKa = 2.95NCINTVNPDD458 pKa = 3.46QLDD461 pKa = 3.95SNNDD465 pKa = 3.21GTGDD469 pKa = 3.41LCEE472 pKa = 4.31QSDD475 pKa = 4.2EE476 pKa = 4.25QVTLIPKK483 pKa = 9.63KK484 pKa = 10.66VNCKK488 pKa = 6.27TLKK491 pKa = 10.71GNVPMTLLGSIDD503 pKa = 4.14FNVSSIDD510 pKa = 4.32ILTLDD515 pKa = 3.53INQVSVAEE523 pKa = 4.09KK524 pKa = 10.42HH525 pKa = 5.9NRR527 pKa = 11.84LHH529 pKa = 7.38LSDD532 pKa = 4.54INNDD536 pKa = 3.36GYY538 pKa = 11.81NDD540 pKa = 4.12AKK542 pKa = 10.69IHH544 pKa = 5.72LNKK547 pKa = 10.34SEE549 pKa = 4.09FCKK552 pKa = 10.86AMDD555 pKa = 4.22TLPAGNPKK563 pKa = 10.21EE564 pKa = 4.51FILNGQFGDD573 pKa = 3.69PSSSFEE579 pKa = 4.02STNSVFIKK587 pKa = 10.53KK588 pKa = 9.95
MM1 pKa = 7.59SRR3 pKa = 11.84FSPVNFSKK11 pKa = 11.09YY12 pKa = 10.58LLLILTTSAVLFSTPSIAITISDD35 pKa = 3.51VSTTNFSNSDD45 pKa = 3.3HH46 pKa = 6.31YY47 pKa = 11.5QDD49 pKa = 5.55DD50 pKa = 4.24GNSLYY55 pKa = 10.6RR56 pKa = 11.84EE57 pKa = 4.06YY58 pKa = 11.24SSNLDD63 pKa = 2.86ITTVTNNNNDD73 pKa = 4.01AIINGHH79 pKa = 6.19LAWFHH84 pKa = 5.6GMRR87 pKa = 11.84VDD89 pKa = 4.36QPEE92 pKa = 4.54GPNFALIYY100 pKa = 9.0RR101 pKa = 11.84HH102 pKa = 6.13NIGYY106 pKa = 9.18EE107 pKa = 3.48ITFTVNDD114 pKa = 5.12PIEE117 pKa = 4.3EE118 pKa = 4.73GYY120 pKa = 10.46EE121 pKa = 3.73ISIEE125 pKa = 3.91QHH127 pKa = 4.91MKK129 pKa = 10.91GLITVSRR136 pKa = 11.84EE137 pKa = 3.53EE138 pKa = 4.73AISVSATAGLMLGRR152 pKa = 11.84VDD154 pKa = 3.57EE155 pKa = 4.62HH156 pKa = 8.14QGDD159 pKa = 4.53GPIHH163 pKa = 6.36LAGFSISGGGLSVDD177 pKa = 4.12SEE179 pKa = 4.49ATDD182 pKa = 3.65TVQTITIDD190 pKa = 3.49EE191 pKa = 4.59TDD193 pKa = 3.65TQNLATQYY201 pKa = 10.58IGTHH205 pKa = 5.24TFTMSFSSFPSPALVSIFQNSGGGEE230 pKa = 3.84TSIQFGVTSNQDD242 pKa = 2.59AFLYY246 pKa = 11.01GNATQNGLIDD256 pKa = 4.48LTPLGHH262 pKa = 6.9FSQINITSLNLTAPDD277 pKa = 3.68TDD279 pKa = 3.6NDD281 pKa = 4.01GVADD285 pKa = 4.6NIDD288 pKa = 3.65NCVLTSNPDD297 pKa = 3.41QLDD300 pKa = 3.49TDD302 pKa = 4.08GDD304 pKa = 4.5GVGDD308 pKa = 3.69VCDD311 pKa = 3.66NCMEE315 pKa = 4.54TPNSDD320 pKa = 4.37QLDD323 pKa = 3.74TDD325 pKa = 3.49EE326 pKa = 6.42DD327 pKa = 5.07GIGDD331 pKa = 3.77VCDD334 pKa = 3.7NCLDD338 pKa = 3.86TSNPDD343 pKa = 3.28QLDD346 pKa = 3.2IDD348 pKa = 4.0EE349 pKa = 5.64DD350 pKa = 4.94GIGDD354 pKa = 3.77VCDD357 pKa = 3.47NCMDD361 pKa = 4.23TSNPDD366 pKa = 3.2QLDD369 pKa = 3.2IDD371 pKa = 4.0EE372 pKa = 5.64DD373 pKa = 4.94GIGDD377 pKa = 3.77VCDD380 pKa = 3.48NCMDD384 pKa = 4.28TPNSDD389 pKa = 3.56QLDD392 pKa = 3.5IDD394 pKa = 3.78EE395 pKa = 5.67DD396 pKa = 4.94GIGDD400 pKa = 3.74VCDD403 pKa = 3.48NCMGTPNSDD412 pKa = 3.91QLDD415 pKa = 3.74TDD417 pKa = 3.6EE418 pKa = 6.34DD419 pKa = 4.72GVGDD423 pKa = 3.82VCDD426 pKa = 3.83NCVDD430 pKa = 3.8TSNPDD435 pKa = 3.54QVDD438 pKa = 3.43TDD440 pKa = 4.14DD441 pKa = 6.01DD442 pKa = 5.44GIGDD446 pKa = 3.75MCDD449 pKa = 2.95NCINTVNPDD458 pKa = 3.46QLDD461 pKa = 3.95SNNDD465 pKa = 3.21GTGDD469 pKa = 3.41LCEE472 pKa = 4.31QSDD475 pKa = 4.2EE476 pKa = 4.25QVTLIPKK483 pKa = 9.63KK484 pKa = 10.66VNCKK488 pKa = 6.27TLKK491 pKa = 10.71GNVPMTLLGSIDD503 pKa = 4.14FNVSSIDD510 pKa = 4.32ILTLDD515 pKa = 3.53INQVSVAEE523 pKa = 4.09KK524 pKa = 10.42HH525 pKa = 5.9NRR527 pKa = 11.84LHH529 pKa = 7.38LSDD532 pKa = 4.54INNDD536 pKa = 3.36GYY538 pKa = 11.81NDD540 pKa = 4.12AKK542 pKa = 10.69IHH544 pKa = 5.72LNKK547 pKa = 10.34SEE549 pKa = 4.09FCKK552 pKa = 10.86AMDD555 pKa = 4.22TLPAGNPKK563 pKa = 10.21EE564 pKa = 4.51FILNGQFGDD573 pKa = 3.69PSSSFEE579 pKa = 4.02STNSVFIKK587 pKa = 10.53KK588 pKa = 9.95
Molecular weight: 63.34 kDa
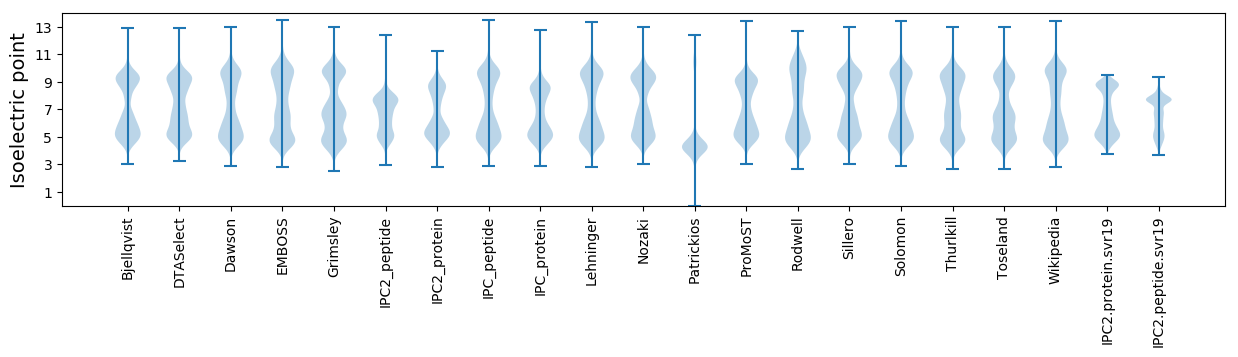
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I1JWV7|A0A1I1JWV7_9GAMM Putative redox protein OS=Pseudoalteromonas denitrificans DSM 6059 OX=1123010 GN=SAMN02745724_01875 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.36RR14 pKa = 11.84SHH16 pKa = 6.15GFRR19 pKa = 11.84ARR21 pKa = 11.84MATTNGRR28 pKa = 11.84KK29 pKa = 9.47LINRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.79GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.42RR12 pKa = 11.84KK13 pKa = 9.36RR14 pKa = 11.84SHH16 pKa = 6.15GFRR19 pKa = 11.84ARR21 pKa = 11.84MATTNGRR28 pKa = 11.84KK29 pKa = 9.47LINRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.79GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
Molecular weight: 5.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1773304 |
39 |
4786 |
332.6 |
37.25 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.341 ± 0.038 | 1.017 ± 0.011 |
5.482 ± 0.03 | 5.896 ± 0.033 |
4.529 ± 0.022 | 6.011 ± 0.034 |
2.16 ± 0.018 | 7.538 ± 0.025 |
6.902 ± 0.036 | 10.46 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.307 ± 0.014 | 5.68 ± 0.032 |
3.425 ± 0.017 | 4.549 ± 0.031 |
3.528 ± 0.023 | 7.205 ± 0.031 |
5.51 ± 0.033 | 6.037 ± 0.03 |
1.121 ± 0.011 | 3.3 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
