
Dokdonella fugitiva
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Xanthomonadales; Rhodanobacteraceae; Dokdonella
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
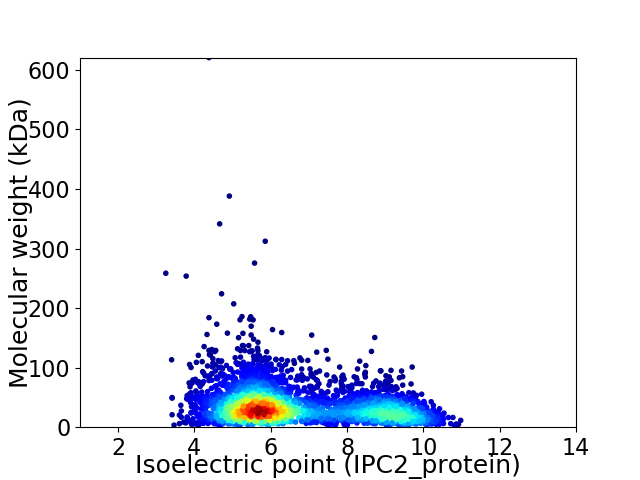
Virtual 2D-PAGE plot for 3614 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R2HXH2|A0A4R2HXH2_9GAMM FG-GAP repeat protein OS=Dokdonella fugitiva OX=328517 GN=EV148_11343 PE=4 SV=1
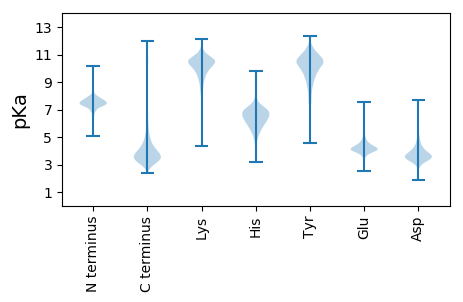
MM1 pKa = 7.29NCISKK6 pKa = 10.82ANAKK10 pKa = 10.01LPSATVLTCAVGVVLAMASSIAHH33 pKa = 6.69AEE35 pKa = 4.16MFCVDD40 pKa = 3.86ATAGLQIALDD50 pKa = 4.08DD51 pKa = 4.53ALSNAEE57 pKa = 3.94PDD59 pKa = 3.91EE60 pKa = 4.24IRR62 pKa = 11.84IQAGDD67 pKa = 3.99YY68 pKa = 10.02ILSAGLVYY76 pKa = 9.6NTQVPGADD84 pKa = 3.11QHH86 pKa = 6.96ALTISGGYY94 pKa = 9.89DD95 pKa = 3.27ADD97 pKa = 3.72CVQRR101 pKa = 11.84IGVSKK106 pKa = 11.04LNGNTQVQVIKK117 pKa = 10.46LVLGAPVFMDD127 pKa = 3.65HH128 pKa = 5.38VTIAGGRR135 pKa = 11.84VEE137 pKa = 4.38NTPGDD142 pKa = 3.79SGRR145 pKa = 11.84GGGVFAQLQGSSAEE159 pKa = 3.96LHH161 pKa = 6.36LDD163 pKa = 3.31AVRR166 pKa = 11.84LISNVASGFGGGIAVISNVGGGSHH190 pKa = 7.2FSLRR194 pKa = 11.84NSLLAGNVAQGAGGAAIGVVSKK216 pKa = 11.05GMVDD220 pKa = 3.23ISNNTITANVSDD232 pKa = 4.22NASAVHH238 pKa = 6.08LQANDD243 pKa = 3.06ASAQFRR249 pKa = 11.84VDD251 pKa = 4.15NNIVWGNGASPSYY264 pKa = 10.49DD265 pKa = 3.52LQLVAPPAGGDD276 pKa = 3.66PYY278 pKa = 11.24HH279 pKa = 6.95LVANDD284 pKa = 3.13IGSTQGMPGDD294 pKa = 4.69DD295 pKa = 3.45SDD297 pKa = 5.72LNVSIDD303 pKa = 3.8PLFVACAFCSNYY315 pKa = 9.33PLSAASPLIDD325 pKa = 4.83AGVGDD330 pKa = 4.33PAGGLAQTDD339 pKa = 3.96LLGAARR345 pKa = 11.84VMGASVDD352 pKa = 3.31MGAYY356 pKa = 9.9EE357 pKa = 4.23FVQPNIAPTVAEE369 pKa = 4.12DD370 pKa = 3.22QAFVVDD376 pKa = 5.03EE377 pKa = 4.53SAAQGAIVGTVTATDD392 pKa = 4.09DD393 pKa = 4.02YY394 pKa = 11.24LPQPHH399 pKa = 7.79AITYY403 pKa = 9.96AIVDD407 pKa = 4.33GNTGDD412 pKa = 4.55AFAIDD417 pKa = 4.17ADD419 pKa = 4.26SGVLTVANPDD429 pKa = 3.94ALDD432 pKa = 3.7STAMPAYY439 pKa = 10.09ALAIKK444 pKa = 9.17VTDD447 pKa = 4.02GEE449 pKa = 4.36LDD451 pKa = 3.45GVGVVNVTVSGPGEE465 pKa = 4.04SDD467 pKa = 4.49VIFQDD472 pKa = 4.23GFDD475 pKa = 3.58GG476 pKa = 4.7
MM1 pKa = 7.29NCISKK6 pKa = 10.82ANAKK10 pKa = 10.01LPSATVLTCAVGVVLAMASSIAHH33 pKa = 6.69AEE35 pKa = 4.16MFCVDD40 pKa = 3.86ATAGLQIALDD50 pKa = 4.08DD51 pKa = 4.53ALSNAEE57 pKa = 3.94PDD59 pKa = 3.91EE60 pKa = 4.24IRR62 pKa = 11.84IQAGDD67 pKa = 3.99YY68 pKa = 10.02ILSAGLVYY76 pKa = 9.6NTQVPGADD84 pKa = 3.11QHH86 pKa = 6.96ALTISGGYY94 pKa = 9.89DD95 pKa = 3.27ADD97 pKa = 3.72CVQRR101 pKa = 11.84IGVSKK106 pKa = 11.04LNGNTQVQVIKK117 pKa = 10.46LVLGAPVFMDD127 pKa = 3.65HH128 pKa = 5.38VTIAGGRR135 pKa = 11.84VEE137 pKa = 4.38NTPGDD142 pKa = 3.79SGRR145 pKa = 11.84GGGVFAQLQGSSAEE159 pKa = 3.96LHH161 pKa = 6.36LDD163 pKa = 3.31AVRR166 pKa = 11.84LISNVASGFGGGIAVISNVGGGSHH190 pKa = 7.2FSLRR194 pKa = 11.84NSLLAGNVAQGAGGAAIGVVSKK216 pKa = 11.05GMVDD220 pKa = 3.23ISNNTITANVSDD232 pKa = 4.22NASAVHH238 pKa = 6.08LQANDD243 pKa = 3.06ASAQFRR249 pKa = 11.84VDD251 pKa = 4.15NNIVWGNGASPSYY264 pKa = 10.49DD265 pKa = 3.52LQLVAPPAGGDD276 pKa = 3.66PYY278 pKa = 11.24HH279 pKa = 6.95LVANDD284 pKa = 3.13IGSTQGMPGDD294 pKa = 4.69DD295 pKa = 3.45SDD297 pKa = 5.72LNVSIDD303 pKa = 3.8PLFVACAFCSNYY315 pKa = 9.33PLSAASPLIDD325 pKa = 4.83AGVGDD330 pKa = 4.33PAGGLAQTDD339 pKa = 3.96LLGAARR345 pKa = 11.84VMGASVDD352 pKa = 3.31MGAYY356 pKa = 9.9EE357 pKa = 4.23FVQPNIAPTVAEE369 pKa = 4.12DD370 pKa = 3.22QAFVVDD376 pKa = 5.03EE377 pKa = 4.53SAAQGAIVGTVTATDD392 pKa = 4.09DD393 pKa = 4.02YY394 pKa = 11.24LPQPHH399 pKa = 7.79AITYY403 pKa = 9.96AIVDD407 pKa = 4.33GNTGDD412 pKa = 4.55AFAIDD417 pKa = 4.17ADD419 pKa = 4.26SGVLTVANPDD429 pKa = 3.94ALDD432 pKa = 3.7STAMPAYY439 pKa = 10.09ALAIKK444 pKa = 9.17VTDD447 pKa = 4.02GEE449 pKa = 4.36LDD451 pKa = 3.45GVGVVNVTVSGPGEE465 pKa = 4.04SDD467 pKa = 4.49VIFQDD472 pKa = 4.23GFDD475 pKa = 3.58GG476 pKa = 4.7
Molecular weight: 47.83 kDa
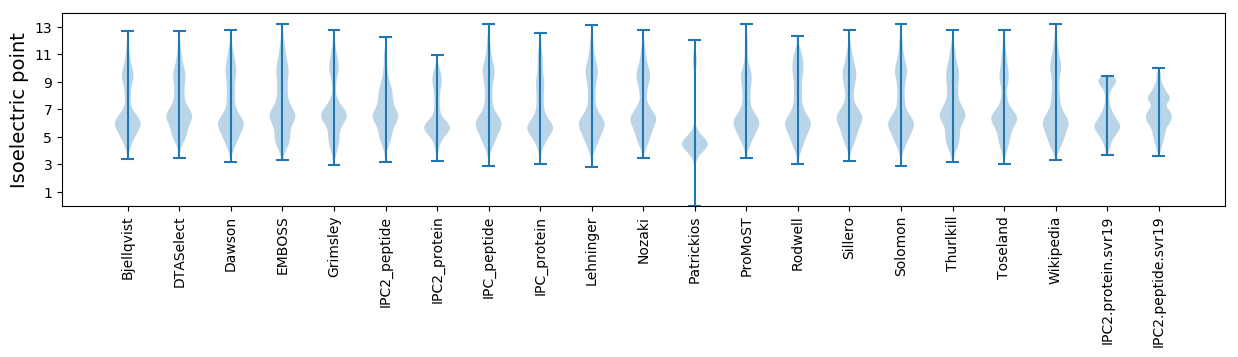
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R2IEM7|A0A4R2IEM7_9GAMM WbqC-like protein OS=Dokdonella fugitiva OX=328517 GN=EV148_10147 PE=4 SV=1
MM1 pKa = 7.06FVAPLSLPSAATRR14 pKa = 11.84AKK16 pKa = 10.18PIAHH20 pKa = 6.4SAWAARR26 pKa = 11.84SPIPWLAGWLLAGLVAVAFIPALRR50 pKa = 11.84GGSTFGATVPFWLVVAPAVDD70 pKa = 4.29LAWLARR76 pKa = 11.84ARR78 pKa = 11.84IGAATSRR85 pKa = 11.84LLRR88 pKa = 11.84RR89 pKa = 11.84MLQRR93 pKa = 11.84TPGQARR99 pKa = 11.84RR100 pKa = 11.84LTVRR104 pKa = 11.84AKK106 pKa = 10.33RR107 pKa = 3.58
MM1 pKa = 7.06FVAPLSLPSAATRR14 pKa = 11.84AKK16 pKa = 10.18PIAHH20 pKa = 6.4SAWAARR26 pKa = 11.84SPIPWLAGWLLAGLVAVAFIPALRR50 pKa = 11.84GGSTFGATVPFWLVVAPAVDD70 pKa = 4.29LAWLARR76 pKa = 11.84ARR78 pKa = 11.84IGAATSRR85 pKa = 11.84LLRR88 pKa = 11.84RR89 pKa = 11.84MLQRR93 pKa = 11.84TPGQARR99 pKa = 11.84RR100 pKa = 11.84LTVRR104 pKa = 11.84AKK106 pKa = 10.33RR107 pKa = 3.58
Molecular weight: 11.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1270453 |
26 |
5886 |
351.5 |
37.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.483 ± 0.066 | 0.927 ± 0.015 |
6.182 ± 0.034 | 5.065 ± 0.043 |
3.383 ± 0.025 | 8.797 ± 0.049 |
2.306 ± 0.022 | 4.04 ± 0.026 |
2.377 ± 0.033 | 10.369 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.836 ± 0.017 | 2.409 ± 0.031 |
5.383 ± 0.03 | 2.979 ± 0.025 |
8.028 ± 0.06 | 4.982 ± 0.038 |
5.096 ± 0.058 | 7.566 ± 0.03 |
1.493 ± 0.018 | 2.301 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
