
Flavobacterium sangjuense
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Flavobacterium
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
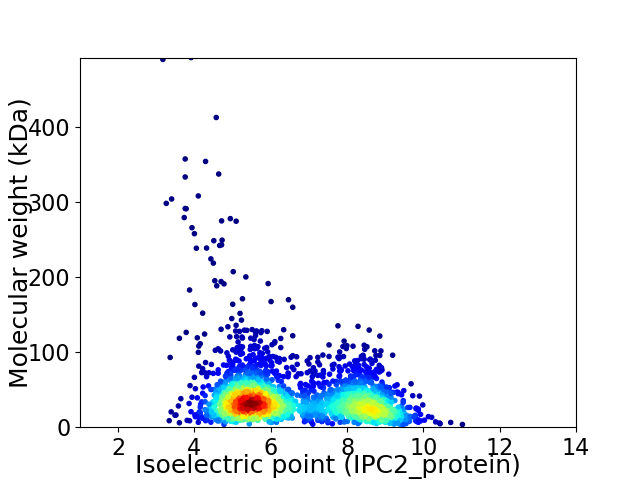
Virtual 2D-PAGE plot for 2637 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
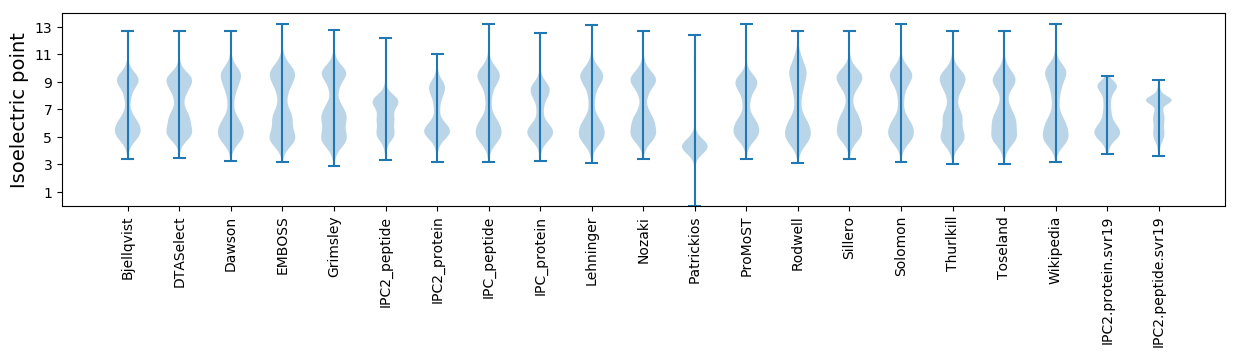
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P7PSQ4|A0A4P7PSQ4_9FLAO Uncharacterized protein OS=Flavobacterium sangjuense OX=2518177 GN=GS03_01174 PE=4 SV=1
MM1 pKa = 6.41KK2 pKa = 10.16QKK4 pKa = 10.85YY5 pKa = 9.65ILFILFSLFVFKK17 pKa = 11.17GNAQSNYY24 pKa = 9.39AVNSIPFQPYY34 pKa = 10.05SGTLSPLTTADD45 pKa = 4.52DD46 pKa = 3.87IYY48 pKa = 11.37SPVITMPFSFDD59 pKa = 3.71FYY61 pKa = 11.09GNSYY65 pKa = 7.73TQFVVSTNGYY75 pKa = 8.78VDD77 pKa = 4.41FRR79 pKa = 11.84TSLAGQYY86 pKa = 10.43SPWNVVYY93 pKa = 8.37TIPNAGFGTKK103 pKa = 10.23NSILGCYY110 pKa = 9.37EE111 pKa = 4.03DD112 pKa = 5.46LNNNSTGAIGTITSGVYY129 pKa = 8.75GTAPYY134 pKa = 10.43RR135 pKa = 11.84KK136 pKa = 8.74FVVYY140 pKa = 9.52FNNQPHH146 pKa = 5.74FQCNSTVALSSSQIILSEE164 pKa = 4.02TTNIIDD170 pKa = 3.45VQVINRR176 pKa = 11.84TPCLAWQQGRR186 pKa = 11.84GVIGLVNLDD195 pKa = 3.53GTQAIAPPGRR205 pKa = 11.84NTGNWSANQEE215 pKa = 3.3AWRR218 pKa = 11.84FYY220 pKa = 10.92RR221 pKa = 11.84PGYY224 pKa = 8.04YY225 pKa = 9.43PSYY228 pKa = 11.0SFVRR232 pKa = 11.84CDD234 pKa = 3.84DD235 pKa = 4.64DD236 pKa = 4.98SDD238 pKa = 4.57SFVTFDD244 pKa = 3.85LTVAANDD251 pKa = 4.33LSPANPSAISFFEE264 pKa = 4.87DD265 pKa = 3.03NALTIPVANPTAYY278 pKa = 9.7INTSNPRR285 pKa = 11.84TVYY288 pKa = 10.97ASGNGAIRR296 pKa = 11.84SVILSVIDD304 pKa = 3.79CSIDD308 pKa = 3.46ADD310 pKa = 4.01SDD312 pKa = 4.31SVPSATEE319 pKa = 3.77DD320 pKa = 3.57VNNDD324 pKa = 2.97TNLANDD330 pKa = 3.97DD331 pKa = 4.11TDD333 pKa = 5.49LDD335 pKa = 4.78GIPNYY340 pKa = 10.49LDD342 pKa = 4.06NDD344 pKa = 4.18DD345 pKa = 6.02DD346 pKa = 5.88GDD348 pKa = 4.08LVLTSVEE355 pKa = 4.29YY356 pKa = 11.04VFARR360 pKa = 11.84VNPRR364 pKa = 11.84QVNVILDD371 pKa = 3.74TDD373 pKa = 4.19LDD375 pKa = 4.73GIPNYY380 pKa = 10.49LDD382 pKa = 4.14NDD384 pKa = 4.21DD385 pKa = 5.81DD386 pKa = 6.21GDD388 pKa = 4.45GLLTWRR394 pKa = 11.84EE395 pKa = 4.24DD396 pKa = 3.71YY397 pKa = 11.44NHH399 pKa = 7.52DD400 pKa = 4.15GNPGNDD406 pKa = 3.69DD407 pKa = 3.84TNSSGTADD415 pKa = 3.64YY416 pKa = 11.16LEE418 pKa = 4.76SSVALGVTPVSIDD431 pKa = 3.36NNSIKK436 pKa = 10.41VFPNPATNVLNIQNNTDD453 pKa = 3.51DD454 pKa = 4.01TNASIEE460 pKa = 4.12IYY462 pKa = 10.05SISGAKK468 pKa = 9.52VKK470 pKa = 10.42SLKK473 pKa = 7.82TTQALTTIAVSDD485 pKa = 4.16LQSGVYY491 pKa = 8.67FVKK494 pKa = 9.72VTMNNQVGNYY504 pKa = 9.79KK505 pKa = 10.46FIKK508 pKa = 9.93NN509 pKa = 3.51
MM1 pKa = 6.41KK2 pKa = 10.16QKK4 pKa = 10.85YY5 pKa = 9.65ILFILFSLFVFKK17 pKa = 11.17GNAQSNYY24 pKa = 9.39AVNSIPFQPYY34 pKa = 10.05SGTLSPLTTADD45 pKa = 4.52DD46 pKa = 3.87IYY48 pKa = 11.37SPVITMPFSFDD59 pKa = 3.71FYY61 pKa = 11.09GNSYY65 pKa = 7.73TQFVVSTNGYY75 pKa = 8.78VDD77 pKa = 4.41FRR79 pKa = 11.84TSLAGQYY86 pKa = 10.43SPWNVVYY93 pKa = 8.37TIPNAGFGTKK103 pKa = 10.23NSILGCYY110 pKa = 9.37EE111 pKa = 4.03DD112 pKa = 5.46LNNNSTGAIGTITSGVYY129 pKa = 8.75GTAPYY134 pKa = 10.43RR135 pKa = 11.84KK136 pKa = 8.74FVVYY140 pKa = 9.52FNNQPHH146 pKa = 5.74FQCNSTVALSSSQIILSEE164 pKa = 4.02TTNIIDD170 pKa = 3.45VQVINRR176 pKa = 11.84TPCLAWQQGRR186 pKa = 11.84GVIGLVNLDD195 pKa = 3.53GTQAIAPPGRR205 pKa = 11.84NTGNWSANQEE215 pKa = 3.3AWRR218 pKa = 11.84FYY220 pKa = 10.92RR221 pKa = 11.84PGYY224 pKa = 8.04YY225 pKa = 9.43PSYY228 pKa = 11.0SFVRR232 pKa = 11.84CDD234 pKa = 3.84DD235 pKa = 4.64DD236 pKa = 4.98SDD238 pKa = 4.57SFVTFDD244 pKa = 3.85LTVAANDD251 pKa = 4.33LSPANPSAISFFEE264 pKa = 4.87DD265 pKa = 3.03NALTIPVANPTAYY278 pKa = 9.7INTSNPRR285 pKa = 11.84TVYY288 pKa = 10.97ASGNGAIRR296 pKa = 11.84SVILSVIDD304 pKa = 3.79CSIDD308 pKa = 3.46ADD310 pKa = 4.01SDD312 pKa = 4.31SVPSATEE319 pKa = 3.77DD320 pKa = 3.57VNNDD324 pKa = 2.97TNLANDD330 pKa = 3.97DD331 pKa = 4.11TDD333 pKa = 5.49LDD335 pKa = 4.78GIPNYY340 pKa = 10.49LDD342 pKa = 4.06NDD344 pKa = 4.18DD345 pKa = 6.02DD346 pKa = 5.88GDD348 pKa = 4.08LVLTSVEE355 pKa = 4.29YY356 pKa = 11.04VFARR360 pKa = 11.84VNPRR364 pKa = 11.84QVNVILDD371 pKa = 3.74TDD373 pKa = 4.19LDD375 pKa = 4.73GIPNYY380 pKa = 10.49LDD382 pKa = 4.14NDD384 pKa = 4.21DD385 pKa = 5.81DD386 pKa = 6.21GDD388 pKa = 4.45GLLTWRR394 pKa = 11.84EE395 pKa = 4.24DD396 pKa = 3.71YY397 pKa = 11.44NHH399 pKa = 7.52DD400 pKa = 4.15GNPGNDD406 pKa = 3.69DD407 pKa = 3.84TNSSGTADD415 pKa = 3.64YY416 pKa = 11.16LEE418 pKa = 4.76SSVALGVTPVSIDD431 pKa = 3.36NNSIKK436 pKa = 10.41VFPNPATNVLNIQNNTDD453 pKa = 3.51DD454 pKa = 4.01TNASIEE460 pKa = 4.12IYY462 pKa = 10.05SISGAKK468 pKa = 9.52VKK470 pKa = 10.42SLKK473 pKa = 7.82TTQALTTIAVSDD485 pKa = 4.16LQSGVYY491 pKa = 8.67FVKK494 pKa = 9.72VTMNNQVGNYY504 pKa = 9.79KK505 pKa = 10.46FIKK508 pKa = 9.93NN509 pKa = 3.51
Molecular weight: 55.47 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P7PS10|A0A4P7PS10_9FLAO Response regulatory domain-containing protein OS=Flavobacterium sangjuense OX=2518177 GN=GS03_01127 PE=4 SV=1
MM1 pKa = 7.84PSGKK5 pKa = 9.32KK6 pKa = 9.59RR7 pKa = 11.84KK8 pKa = 7.05RR9 pKa = 11.84HH10 pKa = 5.11KK11 pKa = 10.59VATHH15 pKa = 5.35KK16 pKa = 10.31RR17 pKa = 11.84KK18 pKa = 9.82KK19 pKa = 8.65RR20 pKa = 11.84ARR22 pKa = 11.84ANRR25 pKa = 11.84HH26 pKa = 4.91KK27 pKa = 10.59KK28 pKa = 9.82KK29 pKa = 10.51KK30 pKa = 10.04
MM1 pKa = 7.84PSGKK5 pKa = 9.32KK6 pKa = 9.59RR7 pKa = 11.84KK8 pKa = 7.05RR9 pKa = 11.84HH10 pKa = 5.11KK11 pKa = 10.59VATHH15 pKa = 5.35KK16 pKa = 10.31RR17 pKa = 11.84KK18 pKa = 9.82KK19 pKa = 8.65RR20 pKa = 11.84ARR22 pKa = 11.84ANRR25 pKa = 11.84HH26 pKa = 4.91KK27 pKa = 10.59KK28 pKa = 9.82KK29 pKa = 10.51KK30 pKa = 10.04
Molecular weight: 3.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
965529 |
29 |
4907 |
366.1 |
40.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.888 ± 0.065 | 0.909 ± 0.03 |
5.289 ± 0.045 | 5.889 ± 0.087 |
5.233 ± 0.047 | 6.66 ± 0.072 |
1.601 ± 0.028 | 7.838 ± 0.053 |
7.431 ± 0.11 | 8.716 ± 0.07 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.131 ± 0.034 | 6.407 ± 0.049 |
3.542 ± 0.042 | 3.317 ± 0.026 |
2.965 ± 0.041 | 6.689 ± 0.06 |
6.952 ± 0.165 | 6.49 ± 0.039 |
1.02 ± 0.015 | 4.035 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
