
Hubei tombus-like virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.4
Get precalculated fractions of proteins
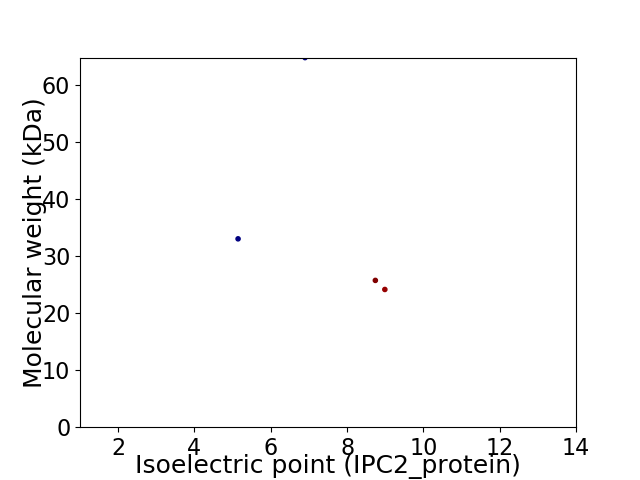
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
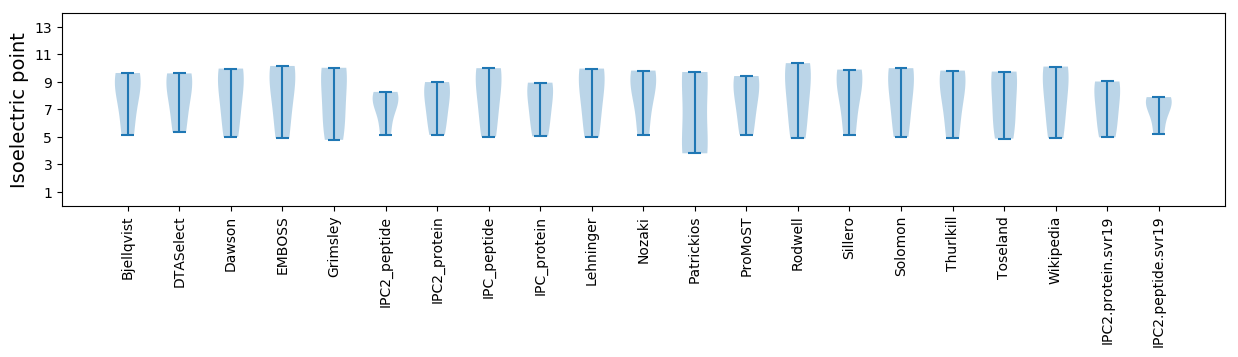
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KH12|A0A1L3KH12_9VIRU Capsid protein OS=Hubei tombus-like virus 2 OX=1923266 PE=3 SV=1
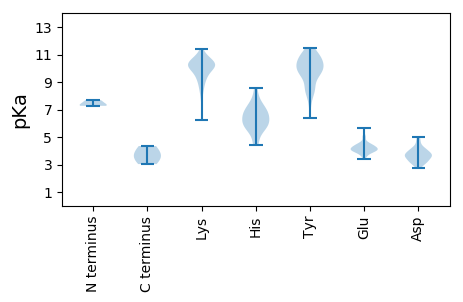
MM1 pKa = 7.27PTSNYY6 pKa = 10.24DD7 pKa = 3.21VLGIGGSTTLRR18 pKa = 11.84AADD21 pKa = 3.68GTQLISYY28 pKa = 9.43DD29 pKa = 3.75PGSAGSIMVNVSGKK43 pKa = 9.59PLFVDD48 pKa = 4.92FTAFFPDD55 pKa = 3.44VRR57 pKa = 11.84KK58 pKa = 10.5SGIGRR63 pKa = 11.84PSLKK67 pKa = 9.25TRR69 pKa = 11.84HH70 pKa = 6.17NLTEE74 pKa = 4.23SYY76 pKa = 8.39QTVVDD81 pKa = 4.55EE82 pKa = 4.57NADD85 pKa = 3.43LSRR88 pKa = 11.84FGGEE92 pKa = 3.63PKK94 pKa = 10.22KK95 pKa = 10.78GYY97 pKa = 9.9EE98 pKa = 4.06YY99 pKa = 9.94STSRR103 pKa = 11.84TIQPDD108 pKa = 4.25GYY110 pKa = 10.89FILQWADD117 pKa = 3.47LNDD120 pKa = 4.05SRR122 pKa = 11.84TMPQTRR128 pKa = 11.84VKK130 pKa = 10.85VVISDD135 pKa = 3.72TLGEE139 pKa = 4.32RR140 pKa = 11.84GPVVTGGSRR149 pKa = 11.84DD150 pKa = 3.71TVDD153 pKa = 3.14GKK155 pKa = 9.83GTEE158 pKa = 4.38VIAITRR164 pKa = 11.84QPTVGASFTSWAGSYY179 pKa = 9.95TYY181 pKa = 10.47TDD183 pKa = 3.59PLGFGANINRR193 pKa = 11.84LPSFITATRR202 pKa = 11.84YY203 pKa = 9.99RR204 pKa = 11.84LGGSMDD210 pKa = 3.61SVALRR215 pKa = 11.84NDD217 pKa = 2.8SDD219 pKa = 3.86MEE221 pKa = 4.18YY222 pKa = 10.41VICFSLVINNFQGSIRR238 pKa = 11.84YY239 pKa = 7.43YY240 pKa = 10.08CPNGEE245 pKa = 4.06RR246 pKa = 11.84TEE248 pKa = 4.09MEE250 pKa = 4.21LNGRR254 pKa = 11.84TIGVATVAFPPGGIMAYY271 pKa = 10.18GYY273 pKa = 10.19DD274 pKa = 3.86AYY276 pKa = 11.01SDD278 pKa = 3.43TGFPPVAVTMSVVPGKK294 pKa = 10.77YY295 pKa = 9.58SDD297 pKa = 3.65VFTTGISVLL306 pKa = 3.8
MM1 pKa = 7.27PTSNYY6 pKa = 10.24DD7 pKa = 3.21VLGIGGSTTLRR18 pKa = 11.84AADD21 pKa = 3.68GTQLISYY28 pKa = 9.43DD29 pKa = 3.75PGSAGSIMVNVSGKK43 pKa = 9.59PLFVDD48 pKa = 4.92FTAFFPDD55 pKa = 3.44VRR57 pKa = 11.84KK58 pKa = 10.5SGIGRR63 pKa = 11.84PSLKK67 pKa = 9.25TRR69 pKa = 11.84HH70 pKa = 6.17NLTEE74 pKa = 4.23SYY76 pKa = 8.39QTVVDD81 pKa = 4.55EE82 pKa = 4.57NADD85 pKa = 3.43LSRR88 pKa = 11.84FGGEE92 pKa = 3.63PKK94 pKa = 10.22KK95 pKa = 10.78GYY97 pKa = 9.9EE98 pKa = 4.06YY99 pKa = 9.94STSRR103 pKa = 11.84TIQPDD108 pKa = 4.25GYY110 pKa = 10.89FILQWADD117 pKa = 3.47LNDD120 pKa = 4.05SRR122 pKa = 11.84TMPQTRR128 pKa = 11.84VKK130 pKa = 10.85VVISDD135 pKa = 3.72TLGEE139 pKa = 4.32RR140 pKa = 11.84GPVVTGGSRR149 pKa = 11.84DD150 pKa = 3.71TVDD153 pKa = 3.14GKK155 pKa = 9.83GTEE158 pKa = 4.38VIAITRR164 pKa = 11.84QPTVGASFTSWAGSYY179 pKa = 9.95TYY181 pKa = 10.47TDD183 pKa = 3.59PLGFGANINRR193 pKa = 11.84LPSFITATRR202 pKa = 11.84YY203 pKa = 9.99RR204 pKa = 11.84LGGSMDD210 pKa = 3.61SVALRR215 pKa = 11.84NDD217 pKa = 2.8SDD219 pKa = 3.86MEE221 pKa = 4.18YY222 pKa = 10.41VICFSLVINNFQGSIRR238 pKa = 11.84YY239 pKa = 7.43YY240 pKa = 10.08CPNGEE245 pKa = 4.06RR246 pKa = 11.84TEE248 pKa = 4.09MEE250 pKa = 4.21LNGRR254 pKa = 11.84TIGVATVAFPPGGIMAYY271 pKa = 10.18GYY273 pKa = 10.19DD274 pKa = 3.86AYY276 pKa = 11.01SDD278 pKa = 3.43TGFPPVAVTMSVVPGKK294 pKa = 10.77YY295 pKa = 9.58SDD297 pKa = 3.65VFTTGISVLL306 pKa = 3.8
Molecular weight: 33.0 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KH12|A0A1L3KH12_9VIRU Capsid protein OS=Hubei tombus-like virus 2 OX=1923266 PE=3 SV=1
MM1 pKa = 7.69PPKK4 pKa = 10.42NNQVSKK10 pKa = 10.94RR11 pKa = 11.84LAARR15 pKa = 11.84KK16 pKa = 9.29APRR19 pKa = 11.84PKK21 pKa = 9.98TSSSGNRR28 pKa = 11.84LVTAPQSRR36 pKa = 11.84AVRR39 pKa = 11.84ATVPSGDD46 pKa = 3.36RR47 pKa = 11.84KK48 pKa = 10.13QICVMTIKK56 pKa = 10.52RR57 pKa = 11.84PAGLTEE63 pKa = 4.01WAQEE67 pKa = 4.22YY68 pKa = 8.5TLHH71 pKa = 7.43PDD73 pKa = 4.07NIPWLEE79 pKa = 4.47GIAPSYY85 pKa = 9.46QRR87 pKa = 11.84WGLKK91 pKa = 8.79GLKK94 pKa = 8.82VWYY97 pKa = 9.13EE98 pKa = 3.84PRR100 pKa = 11.84ATALTPGTVSMAILSDD116 pKa = 4.13FKK118 pKa = 11.39DD119 pKa = 3.73GTPKK123 pKa = 10.55SLQSLTSVKK132 pKa = 10.26GAVRR136 pKa = 11.84GAPWDD141 pKa = 3.94KK142 pKa = 10.27FTLSCPKK149 pKa = 9.75FRR151 pKa = 11.84TYY153 pKa = 10.87EE154 pKa = 4.01YY155 pKa = 10.57VSDD158 pKa = 3.93VASLSGEE165 pKa = 4.05DD166 pKa = 3.77LNNRR170 pKa = 11.84ALGKK174 pKa = 10.13IVVCADD180 pKa = 3.15MDD182 pKa = 4.08DD183 pKa = 4.09SFTVGSIVGRR193 pKa = 11.84IFIEE197 pKa = 4.21YY198 pKa = 10.43SDD200 pKa = 4.38VLLDD204 pKa = 5.02SIDD207 pKa = 3.41PTLQRR212 pKa = 11.84KK213 pKa = 8.14SAATSPGTT221 pKa = 3.53
MM1 pKa = 7.69PPKK4 pKa = 10.42NNQVSKK10 pKa = 10.94RR11 pKa = 11.84LAARR15 pKa = 11.84KK16 pKa = 9.29APRR19 pKa = 11.84PKK21 pKa = 9.98TSSSGNRR28 pKa = 11.84LVTAPQSRR36 pKa = 11.84AVRR39 pKa = 11.84ATVPSGDD46 pKa = 3.36RR47 pKa = 11.84KK48 pKa = 10.13QICVMTIKK56 pKa = 10.52RR57 pKa = 11.84PAGLTEE63 pKa = 4.01WAQEE67 pKa = 4.22YY68 pKa = 8.5TLHH71 pKa = 7.43PDD73 pKa = 4.07NIPWLEE79 pKa = 4.47GIAPSYY85 pKa = 9.46QRR87 pKa = 11.84WGLKK91 pKa = 8.79GLKK94 pKa = 8.82VWYY97 pKa = 9.13EE98 pKa = 3.84PRR100 pKa = 11.84ATALTPGTVSMAILSDD116 pKa = 4.13FKK118 pKa = 11.39DD119 pKa = 3.73GTPKK123 pKa = 10.55SLQSLTSVKK132 pKa = 10.26GAVRR136 pKa = 11.84GAPWDD141 pKa = 3.94KK142 pKa = 10.27FTLSCPKK149 pKa = 9.75FRR151 pKa = 11.84TYY153 pKa = 10.87EE154 pKa = 4.01YY155 pKa = 10.57VSDD158 pKa = 3.93VASLSGEE165 pKa = 4.05DD166 pKa = 3.77LNNRR170 pKa = 11.84ALGKK174 pKa = 10.13IVVCADD180 pKa = 3.15MDD182 pKa = 4.08DD183 pKa = 4.09SFTVGSIVGRR193 pKa = 11.84IFIEE197 pKa = 4.21YY198 pKa = 10.43SDD200 pKa = 4.38VLLDD204 pKa = 5.02SIDD207 pKa = 3.41PTLQRR212 pKa = 11.84KK213 pKa = 8.14SAATSPGTT221 pKa = 3.53
Molecular weight: 24.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1322 |
221 |
570 |
330.5 |
36.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.413 ± 0.922 | 1.815 ± 0.709 |
5.673 ± 0.319 | 5.9 ± 1.224 |
3.782 ± 0.647 | 8.245 ± 0.912 |
1.664 ± 0.586 | 4.085 ± 0.482 |
4.841 ± 0.765 | 7.186 ± 0.64 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.421 ± 0.175 | 3.631 ± 0.244 |
5.522 ± 0.63 | 2.799 ± 0.214 |
7.64 ± 0.881 | 7.413 ± 0.956 |
6.203 ± 1.222 | 7.943 ± 0.667 |
2.118 ± 0.64 | 3.707 ± 0.642 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
