
Paraglaciecola psychrophila 170
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Alteromonadaceae; Paraglaciecola; Paraglaciecola psychrophila
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
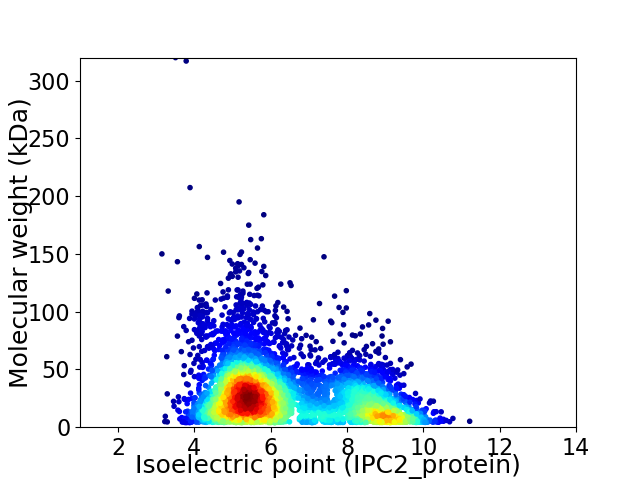
Virtual 2D-PAGE plot for 5601 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K7AW16|K7AW16_9ALTE Shikimate kinase OS=Paraglaciecola psychrophila 170 OX=1129794 GN=aroK PE=3 SV=1
MM1 pKa = 7.76NSFLKK6 pKa = 10.18TGITSALFFICGFAVAAKK24 pKa = 10.21SPLIPYY30 pKa = 10.16SKK32 pKa = 9.47TNSQIAEE39 pKa = 3.92TRR41 pKa = 11.84IVGGEE46 pKa = 3.77EE47 pKa = 4.0AIQEE51 pKa = 3.93NWPWMTAYY59 pKa = 10.65VGVFTNFLTSLSVNNVIYY77 pKa = 7.37EE78 pKa = 4.3TRR80 pKa = 11.84SFTSGVGGQVSGEE93 pKa = 4.05IIACGIGDD101 pKa = 4.77AVCVDD106 pKa = 3.36ATGNVCLIEE115 pKa = 4.49RR116 pKa = 11.84GIVDD120 pKa = 4.44FSEE123 pKa = 5.48KK124 pKa = 10.45SDD126 pKa = 3.32NCEE129 pKa = 3.66AGGGIGAIIYY139 pKa = 10.6NNEE142 pKa = 3.77EE143 pKa = 3.55VDD145 pKa = 4.0NITGGTLGEE154 pKa = 5.32DD155 pKa = 3.64YY156 pKa = 10.51TGTIPIVAVNQKK168 pKa = 10.8DD169 pKa = 3.66GLALLEE175 pKa = 4.13QIGNIASLAVSEE187 pKa = 4.44TTEE190 pKa = 4.05LQQDD194 pKa = 4.26ASCGASFLGDD204 pKa = 3.1KK205 pKa = 9.65WVLTAAHH212 pKa = 6.55CVDD215 pKa = 4.84SEE217 pKa = 4.22NANQFKK223 pKa = 10.53MNVGEE228 pKa = 4.12YY229 pKa = 10.17DD230 pKa = 3.52LSNGAEE236 pKa = 4.02NAIDD240 pKa = 3.3IANIYY245 pKa = 9.67IHH247 pKa = 6.39PQYY250 pKa = 11.06DD251 pKa = 3.19ADD253 pKa = 5.33AISNDD258 pKa = 2.86IALIEE263 pKa = 4.32LASSSDD269 pKa = 3.41AVGVKK274 pKa = 9.85IADD277 pKa = 3.84PDD279 pKa = 3.72VTDD282 pKa = 3.7QYY284 pKa = 11.78AIEE287 pKa = 4.41NSLATVAGWGGRR299 pKa = 11.84AGYY302 pKa = 10.07APNEE306 pKa = 4.42GPTSDD311 pKa = 5.08FPDD314 pKa = 3.72ILHH317 pKa = 6.64KK318 pKa = 10.69VDD320 pKa = 5.98LRR322 pKa = 11.84LTTNAQCRR330 pKa = 11.84EE331 pKa = 4.01EE332 pKa = 4.39LGEE335 pKa = 4.19SFGISAANVGVTDD348 pKa = 4.09VMICAAIPEE357 pKa = 4.62GGRR360 pKa = 11.84GSCQGDD366 pKa = 3.06SGGPLVVNTGSGVQQVGIVSWGFGCAASGYY396 pKa = 8.09PGVYY400 pKa = 9.17TRR402 pKa = 11.84VSEE405 pKa = 4.35FKK407 pKa = 10.51DD408 pKa = 3.4WISAITDD415 pKa = 4.82GIAITQRR422 pKa = 11.84HH423 pKa = 5.09NFGLGLEE430 pKa = 4.84GEE432 pKa = 4.59VQTTGLAVTNNSEE445 pKa = 4.54TNVGLSFAFSDD456 pKa = 4.2SNTFTLDD463 pKa = 3.11ASNCPTLDD471 pKa = 3.7AGNSCQVSVSYY482 pKa = 11.01LPTIANNVSSKK493 pKa = 11.08LIITTDD499 pKa = 3.52DD500 pKa = 3.76PQVQSSSATVTAGTTLGYY518 pKa = 10.43AVEE521 pKa = 4.45LAATAGSEE529 pKa = 4.12SDD531 pKa = 3.46AVTLFSGGTNGASAWVASSTEE552 pKa = 3.92VGIEE556 pKa = 3.95STTTGDD562 pKa = 3.52LQDD565 pKa = 4.53SIFVASIEE573 pKa = 4.33GEE575 pKa = 4.36GVLTFDD581 pKa = 3.53WAVSSEE587 pKa = 3.86ANTLVEE593 pKa = 4.15TDD595 pKa = 3.42PDD597 pKa = 3.91FEE599 pKa = 5.36RR600 pKa = 11.84FDD602 pKa = 4.86ALFLYY607 pKa = 10.98LNGDD611 pKa = 3.71LVDD614 pKa = 4.66VINGNDD620 pKa = 3.59DD621 pKa = 4.02EE622 pKa = 5.38ISFTEE627 pKa = 3.95YY628 pKa = 11.11SLNLSEE634 pKa = 6.52GINLVNWTYY643 pKa = 11.61SKK645 pKa = 11.3DD646 pKa = 3.64PDD648 pKa = 3.86VSEE651 pKa = 5.27GDD653 pKa = 3.34DD654 pKa = 3.2KK655 pKa = 11.92GFIRR659 pKa = 11.84NLVFTATVVAPPPVAVVPIPAPPNSSGGAGSLGWILLCLVGLIFRR704 pKa = 11.84LRR706 pKa = 11.84TKK708 pKa = 10.45HH709 pKa = 5.98
MM1 pKa = 7.76NSFLKK6 pKa = 10.18TGITSALFFICGFAVAAKK24 pKa = 10.21SPLIPYY30 pKa = 10.16SKK32 pKa = 9.47TNSQIAEE39 pKa = 3.92TRR41 pKa = 11.84IVGGEE46 pKa = 3.77EE47 pKa = 4.0AIQEE51 pKa = 3.93NWPWMTAYY59 pKa = 10.65VGVFTNFLTSLSVNNVIYY77 pKa = 7.37EE78 pKa = 4.3TRR80 pKa = 11.84SFTSGVGGQVSGEE93 pKa = 4.05IIACGIGDD101 pKa = 4.77AVCVDD106 pKa = 3.36ATGNVCLIEE115 pKa = 4.49RR116 pKa = 11.84GIVDD120 pKa = 4.44FSEE123 pKa = 5.48KK124 pKa = 10.45SDD126 pKa = 3.32NCEE129 pKa = 3.66AGGGIGAIIYY139 pKa = 10.6NNEE142 pKa = 3.77EE143 pKa = 3.55VDD145 pKa = 4.0NITGGTLGEE154 pKa = 5.32DD155 pKa = 3.64YY156 pKa = 10.51TGTIPIVAVNQKK168 pKa = 10.8DD169 pKa = 3.66GLALLEE175 pKa = 4.13QIGNIASLAVSEE187 pKa = 4.44TTEE190 pKa = 4.05LQQDD194 pKa = 4.26ASCGASFLGDD204 pKa = 3.1KK205 pKa = 9.65WVLTAAHH212 pKa = 6.55CVDD215 pKa = 4.84SEE217 pKa = 4.22NANQFKK223 pKa = 10.53MNVGEE228 pKa = 4.12YY229 pKa = 10.17DD230 pKa = 3.52LSNGAEE236 pKa = 4.02NAIDD240 pKa = 3.3IANIYY245 pKa = 9.67IHH247 pKa = 6.39PQYY250 pKa = 11.06DD251 pKa = 3.19ADD253 pKa = 5.33AISNDD258 pKa = 2.86IALIEE263 pKa = 4.32LASSSDD269 pKa = 3.41AVGVKK274 pKa = 9.85IADD277 pKa = 3.84PDD279 pKa = 3.72VTDD282 pKa = 3.7QYY284 pKa = 11.78AIEE287 pKa = 4.41NSLATVAGWGGRR299 pKa = 11.84AGYY302 pKa = 10.07APNEE306 pKa = 4.42GPTSDD311 pKa = 5.08FPDD314 pKa = 3.72ILHH317 pKa = 6.64KK318 pKa = 10.69VDD320 pKa = 5.98LRR322 pKa = 11.84LTTNAQCRR330 pKa = 11.84EE331 pKa = 4.01EE332 pKa = 4.39LGEE335 pKa = 4.19SFGISAANVGVTDD348 pKa = 4.09VMICAAIPEE357 pKa = 4.62GGRR360 pKa = 11.84GSCQGDD366 pKa = 3.06SGGPLVVNTGSGVQQVGIVSWGFGCAASGYY396 pKa = 8.09PGVYY400 pKa = 9.17TRR402 pKa = 11.84VSEE405 pKa = 4.35FKK407 pKa = 10.51DD408 pKa = 3.4WISAITDD415 pKa = 4.82GIAITQRR422 pKa = 11.84HH423 pKa = 5.09NFGLGLEE430 pKa = 4.84GEE432 pKa = 4.59VQTTGLAVTNNSEE445 pKa = 4.54TNVGLSFAFSDD456 pKa = 4.2SNTFTLDD463 pKa = 3.11ASNCPTLDD471 pKa = 3.7AGNSCQVSVSYY482 pKa = 11.01LPTIANNVSSKK493 pKa = 11.08LIITTDD499 pKa = 3.52DD500 pKa = 3.76PQVQSSSATVTAGTTLGYY518 pKa = 10.43AVEE521 pKa = 4.45LAATAGSEE529 pKa = 4.12SDD531 pKa = 3.46AVTLFSGGTNGASAWVASSTEE552 pKa = 3.92VGIEE556 pKa = 3.95STTTGDD562 pKa = 3.52LQDD565 pKa = 4.53SIFVASIEE573 pKa = 4.33GEE575 pKa = 4.36GVLTFDD581 pKa = 3.53WAVSSEE587 pKa = 3.86ANTLVEE593 pKa = 4.15TDD595 pKa = 3.42PDD597 pKa = 3.91FEE599 pKa = 5.36RR600 pKa = 11.84FDD602 pKa = 4.86ALFLYY607 pKa = 10.98LNGDD611 pKa = 3.71LVDD614 pKa = 4.66VINGNDD620 pKa = 3.59DD621 pKa = 4.02EE622 pKa = 5.38ISFTEE627 pKa = 3.95YY628 pKa = 11.11SLNLSEE634 pKa = 6.52GINLVNWTYY643 pKa = 11.61SKK645 pKa = 11.3DD646 pKa = 3.64PDD648 pKa = 3.86VSEE651 pKa = 5.27GDD653 pKa = 3.34DD654 pKa = 3.2KK655 pKa = 11.92GFIRR659 pKa = 11.84NLVFTATVVAPPPVAVVPIPAPPNSSGGAGSLGWILLCLVGLIFRR704 pKa = 11.84LRR706 pKa = 11.84TKK708 pKa = 10.45HH709 pKa = 5.98
Molecular weight: 73.97 kDa
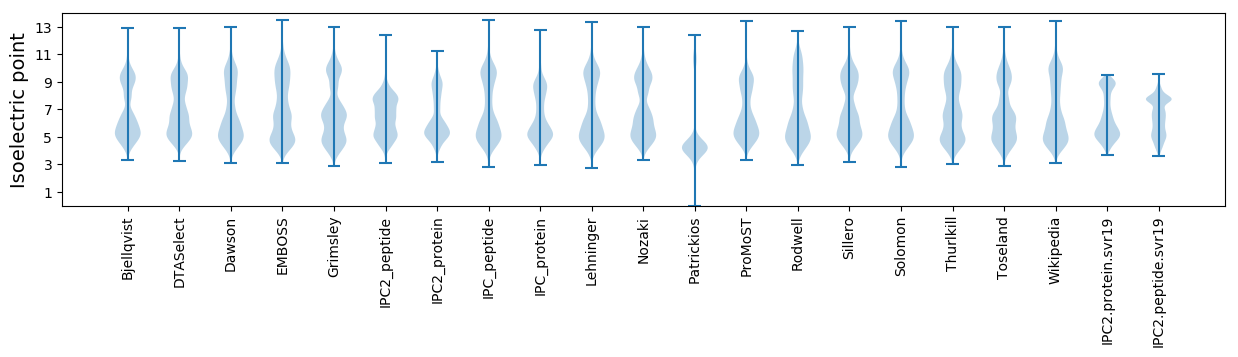
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K7ALB5|K7ALB5_9ALTE Uncharacterized protein OS=Paraglaciecola psychrophila 170 OX=1129794 GN=C427_2761 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.14RR12 pKa = 11.84KK13 pKa = 9.13RR14 pKa = 11.84SHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.38NGRR28 pKa = 11.84KK29 pKa = 9.48VIANRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.35GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.14RR12 pKa = 11.84KK13 pKa = 9.13RR14 pKa = 11.84SHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.38NGRR28 pKa = 11.84KK29 pKa = 9.48VIANRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.35GRR39 pKa = 11.84ARR41 pKa = 11.84LSAA44 pKa = 3.91
Molecular weight: 5.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1496535 |
37 |
3094 |
267.2 |
29.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.108 ± 0.036 | 1.026 ± 0.012 |
5.608 ± 0.035 | 5.711 ± 0.037 |
4.4 ± 0.028 | 6.632 ± 0.042 |
2.207 ± 0.019 | 6.878 ± 0.026 |
5.629 ± 0.035 | 10.344 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.568 ± 0.021 | 4.759 ± 0.03 |
3.75 ± 0.023 | 4.761 ± 0.042 |
4.11 ± 0.028 | 6.957 ± 0.032 |
5.474 ± 0.037 | 6.701 ± 0.032 |
1.236 ± 0.012 | 3.139 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
