
Cryobacterium roopkundense
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Cryobacterium
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
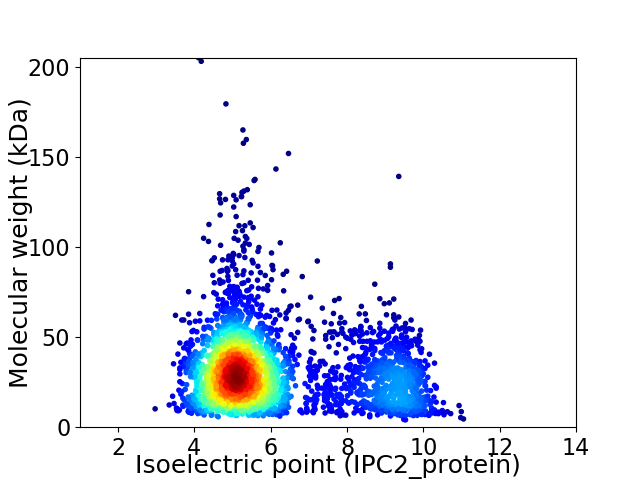
Virtual 2D-PAGE plot for 3249 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A099J456|A0A099J456_9MICO Enoyl-CoA hydratase OS=Cryobacterium roopkundense OX=1001240 GN=BJ997_003394 PE=3 SV=1
MM1 pKa = 6.3EE2 pKa = 4.04TRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84GLMATATVVASALFLTGCVQSDD28 pKa = 3.82RR29 pKa = 11.84DD30 pKa = 3.61DD31 pKa = 3.68TAAGDD36 pKa = 3.6VDD38 pKa = 4.11STFVFAASSDD48 pKa = 3.91PASLDD53 pKa = 3.26PAFASDD59 pKa = 3.96GEE61 pKa = 4.44SFRR64 pKa = 11.84VSRR67 pKa = 11.84QIFEE71 pKa = 4.37GLVGVAPGTADD82 pKa = 3.85PAPLLAEE89 pKa = 4.38SWDD92 pKa = 3.7QSDD95 pKa = 5.27DD96 pKa = 3.56GLSYY100 pKa = 10.85TFALKK105 pKa = 10.63EE106 pKa = 3.95GVTFHH111 pKa = 7.79DD112 pKa = 4.21GTDD115 pKa = 3.81FDD117 pKa = 5.35AEE119 pKa = 4.15AVCFNFDD126 pKa = 2.43RR127 pKa = 11.84WFNFTGIAQSPSVAYY142 pKa = 10.45YY143 pKa = 10.03YY144 pKa = 11.51GNLFRR149 pKa = 11.84AYY151 pKa = 10.35ADD153 pKa = 3.55NPEE156 pKa = 3.95GAVYY160 pKa = 10.12DD161 pKa = 3.91SCSTGDD167 pKa = 3.37TGEE170 pKa = 4.23VTVTLAQPFAGFIAALSLPAFSMQSPTALEE200 pKa = 3.92EE201 pKa = 3.97FAADD205 pKa = 4.32DD206 pKa = 3.45IGGSEE211 pKa = 4.22EE212 pKa = 4.21APALSEE218 pKa = 4.12YY219 pKa = 10.93AQGHH223 pKa = 4.88PTGTGPFAFDD233 pKa = 2.71SWEE236 pKa = 3.89IGSQLEE242 pKa = 3.79LSAYY246 pKa = 9.35SDD248 pKa = 3.65YY249 pKa = 10.74WGEE252 pKa = 3.99QGDD255 pKa = 3.9VQSIIFRR262 pKa = 11.84VIDD265 pKa = 3.94DD266 pKa = 3.78PQARR270 pKa = 11.84RR271 pKa = 11.84QSLEE275 pKa = 3.63AGSIDD280 pKa = 4.58GYY282 pKa = 11.3DD283 pKa = 3.52LVAPADD289 pKa = 4.0TVALEE294 pKa = 4.08TAGFQIVPRR303 pKa = 11.84DD304 pKa = 3.82PFTILYY310 pKa = 10.21LGINQSVPEE319 pKa = 4.17LQDD322 pKa = 3.02VRR324 pKa = 11.84VRR326 pKa = 11.84EE327 pKa = 4.63AISLAIDD334 pKa = 3.32KK335 pKa = 10.25QQLIDD340 pKa = 3.4QTLPEE345 pKa = 4.57GTQLATQFIPPVVAGYY361 pKa = 10.82NDD363 pKa = 4.6DD364 pKa = 3.77VTTYY368 pKa = 10.93DD369 pKa = 3.85YY370 pKa = 11.27DD371 pKa = 3.8QEE373 pKa = 4.2AAKK376 pKa = 10.46QLLADD381 pKa = 4.33AGYY384 pKa = 9.72PDD386 pKa = 5.41GFTLQFNYY394 pKa = 7.61PTGVSRR400 pKa = 11.84PYY402 pKa = 9.69MPTPEE407 pKa = 4.08EE408 pKa = 4.13VFTNISAQLGAIGITIDD425 pKa = 3.76PQPNAWSPDD434 pKa = 3.44YY435 pKa = 10.94LDD437 pKa = 5.69RR438 pKa = 11.84IQGSEE443 pKa = 3.51NHH445 pKa = 6.86GIHH448 pKa = 6.53LLGWTGDD455 pKa = 3.85YY456 pKa = 11.17NDD458 pKa = 3.83TDD460 pKa = 3.59NFVGVFFGTSSLEE473 pKa = 3.94FGFDD477 pKa = 3.43NPEE480 pKa = 3.76LFAALTEE487 pKa = 4.04ARR489 pKa = 11.84GISSLEE495 pKa = 3.95EE496 pKa = 3.65QVPMYY501 pKa = 10.34EE502 pKa = 4.69VINEE506 pKa = 4.73TIAEE510 pKa = 4.9FIPAIPLAHH519 pKa = 7.25PAPSLAFSDD528 pKa = 4.13RR529 pKa = 11.84VEE531 pKa = 4.53SYY533 pKa = 10.33PVSPVNDD540 pKa = 3.39EE541 pKa = 4.12VFNMIVLTKK550 pKa = 10.74
MM1 pKa = 6.3EE2 pKa = 4.04TRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84GLMATATVVASALFLTGCVQSDD28 pKa = 3.82RR29 pKa = 11.84DD30 pKa = 3.61DD31 pKa = 3.68TAAGDD36 pKa = 3.6VDD38 pKa = 4.11STFVFAASSDD48 pKa = 3.91PASLDD53 pKa = 3.26PAFASDD59 pKa = 3.96GEE61 pKa = 4.44SFRR64 pKa = 11.84VSRR67 pKa = 11.84QIFEE71 pKa = 4.37GLVGVAPGTADD82 pKa = 3.85PAPLLAEE89 pKa = 4.38SWDD92 pKa = 3.7QSDD95 pKa = 5.27DD96 pKa = 3.56GLSYY100 pKa = 10.85TFALKK105 pKa = 10.63EE106 pKa = 3.95GVTFHH111 pKa = 7.79DD112 pKa = 4.21GTDD115 pKa = 3.81FDD117 pKa = 5.35AEE119 pKa = 4.15AVCFNFDD126 pKa = 2.43RR127 pKa = 11.84WFNFTGIAQSPSVAYY142 pKa = 10.45YY143 pKa = 10.03YY144 pKa = 11.51GNLFRR149 pKa = 11.84AYY151 pKa = 10.35ADD153 pKa = 3.55NPEE156 pKa = 3.95GAVYY160 pKa = 10.12DD161 pKa = 3.91SCSTGDD167 pKa = 3.37TGEE170 pKa = 4.23VTVTLAQPFAGFIAALSLPAFSMQSPTALEE200 pKa = 3.92EE201 pKa = 3.97FAADD205 pKa = 4.32DD206 pKa = 3.45IGGSEE211 pKa = 4.22EE212 pKa = 4.21APALSEE218 pKa = 4.12YY219 pKa = 10.93AQGHH223 pKa = 4.88PTGTGPFAFDD233 pKa = 2.71SWEE236 pKa = 3.89IGSQLEE242 pKa = 3.79LSAYY246 pKa = 9.35SDD248 pKa = 3.65YY249 pKa = 10.74WGEE252 pKa = 3.99QGDD255 pKa = 3.9VQSIIFRR262 pKa = 11.84VIDD265 pKa = 3.94DD266 pKa = 3.78PQARR270 pKa = 11.84RR271 pKa = 11.84QSLEE275 pKa = 3.63AGSIDD280 pKa = 4.58GYY282 pKa = 11.3DD283 pKa = 3.52LVAPADD289 pKa = 4.0TVALEE294 pKa = 4.08TAGFQIVPRR303 pKa = 11.84DD304 pKa = 3.82PFTILYY310 pKa = 10.21LGINQSVPEE319 pKa = 4.17LQDD322 pKa = 3.02VRR324 pKa = 11.84VRR326 pKa = 11.84EE327 pKa = 4.63AISLAIDD334 pKa = 3.32KK335 pKa = 10.25QQLIDD340 pKa = 3.4QTLPEE345 pKa = 4.57GTQLATQFIPPVVAGYY361 pKa = 10.82NDD363 pKa = 4.6DD364 pKa = 3.77VTTYY368 pKa = 10.93DD369 pKa = 3.85YY370 pKa = 11.27DD371 pKa = 3.8QEE373 pKa = 4.2AAKK376 pKa = 10.46QLLADD381 pKa = 4.33AGYY384 pKa = 9.72PDD386 pKa = 5.41GFTLQFNYY394 pKa = 7.61PTGVSRR400 pKa = 11.84PYY402 pKa = 9.69MPTPEE407 pKa = 4.08EE408 pKa = 4.13VFTNISAQLGAIGITIDD425 pKa = 3.76PQPNAWSPDD434 pKa = 3.44YY435 pKa = 10.94LDD437 pKa = 5.69RR438 pKa = 11.84IQGSEE443 pKa = 3.51NHH445 pKa = 6.86GIHH448 pKa = 6.53LLGWTGDD455 pKa = 3.85YY456 pKa = 11.17NDD458 pKa = 3.83TDD460 pKa = 3.59NFVGVFFGTSSLEE473 pKa = 3.94FGFDD477 pKa = 3.43NPEE480 pKa = 3.76LFAALTEE487 pKa = 4.04ARR489 pKa = 11.84GISSLEE495 pKa = 3.95EE496 pKa = 3.65QVPMYY501 pKa = 10.34EE502 pKa = 4.69VINEE506 pKa = 4.73TIAEE510 pKa = 4.9FIPAIPLAHH519 pKa = 7.25PAPSLAFSDD528 pKa = 4.13RR529 pKa = 11.84VEE531 pKa = 4.53SYY533 pKa = 10.33PVSPVNDD540 pKa = 3.39EE541 pKa = 4.12VFNMIVLTKK550 pKa = 10.74
Molecular weight: 59.45 kDa
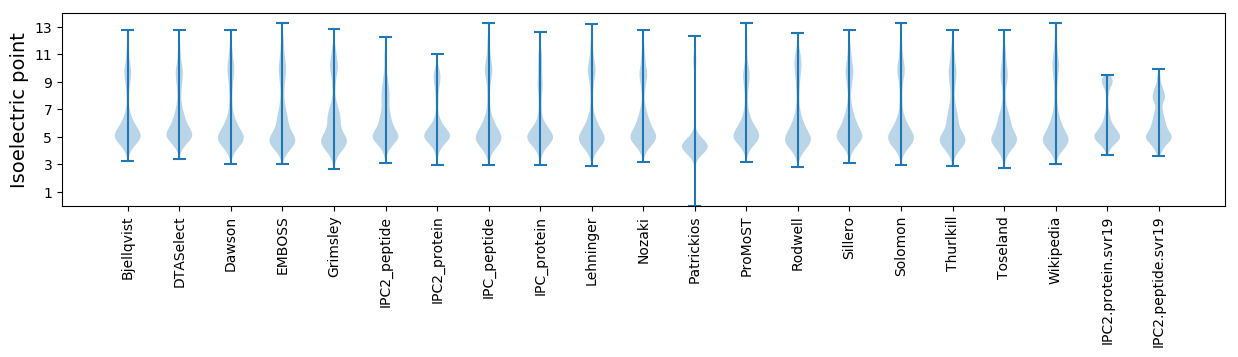
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A099J168|A0A099J168_9MICO DUF4190 domain-containing protein OS=Cryobacterium roopkundense OX=1001240 GN=GY21_18155 PE=4 SV=1
MM1 pKa = 7.65KK2 pKa = 10.33VRR4 pKa = 11.84NSLKK8 pKa = 10.39ALKK11 pKa = 9.59KK12 pKa = 10.37VPGSQIVRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84GRR24 pKa = 11.84TFVINKK30 pKa = 8.63LNPRR34 pKa = 11.84AKK36 pKa = 10.25ARR38 pKa = 11.84QGG40 pKa = 3.19
MM1 pKa = 7.65KK2 pKa = 10.33VRR4 pKa = 11.84NSLKK8 pKa = 10.39ALKK11 pKa = 9.59KK12 pKa = 10.37VPGSQIVRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84GRR24 pKa = 11.84TFVINKK30 pKa = 8.63LNPRR34 pKa = 11.84AKK36 pKa = 10.25ARR38 pKa = 11.84QGG40 pKa = 3.19
Molecular weight: 4.57 kDa
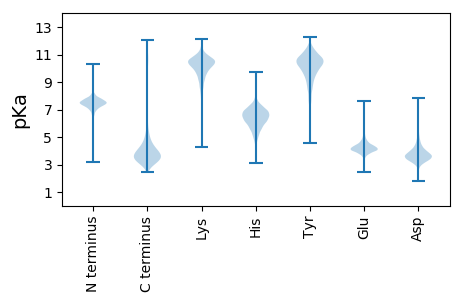
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
988510 |
37 |
1997 |
304.3 |
32.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.836 ± 0.054 | 0.583 ± 0.011 |
5.788 ± 0.031 | 5.303 ± 0.039 |
3.314 ± 0.025 | 8.744 ± 0.039 |
2.015 ± 0.021 | 4.864 ± 0.03 |
2.239 ± 0.032 | 10.447 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.929 ± 0.017 | 2.421 ± 0.023 |
5.106 ± 0.03 | 2.917 ± 0.022 |
6.649 ± 0.04 | 6.117 ± 0.031 |
6.402 ± 0.034 | 8.849 ± 0.042 |
1.423 ± 0.017 | 2.053 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
