
Hortaea werneckii
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; dothideomyceta; Dothideomycetes; Dothideomycetidae; Mycosphaerellales;
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
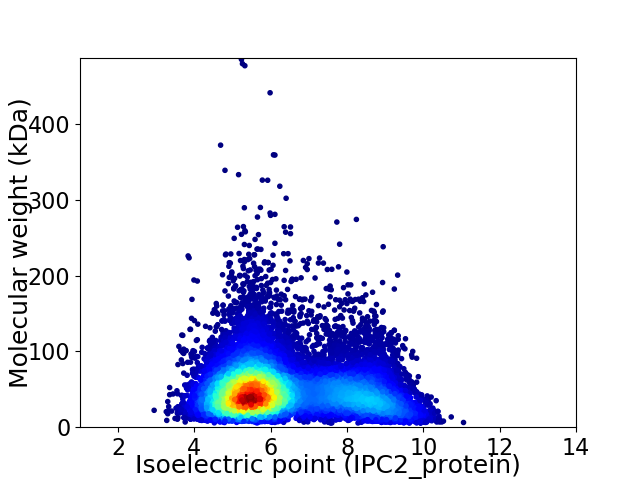
Virtual 2D-PAGE plot for 16466 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A3M6YG17|A0A3M6YG17_HORWE Uncharacterized protein (Fragment) OS=Hortaea werneckii OX=91943 GN=D0866_15707 PE=4 SV=1
MM1 pKa = 7.76APITNFLASSLALLSSTALAANPIKK26 pKa = 10.73VQEE29 pKa = 4.13QEE31 pKa = 4.6FIDD34 pKa = 3.92TKK36 pKa = 9.1TDD38 pKa = 2.94EE39 pKa = 4.34RR40 pKa = 11.84FVIVGVDD47 pKa = 3.48YY48 pKa = 11.34QPGGSAAVGTGNGDD62 pKa = 3.77PLSNADD68 pKa = 3.26EE69 pKa = 4.41CRR71 pKa = 11.84RR72 pKa = 11.84DD73 pKa = 3.39AALMQNMGVNTIRR86 pKa = 11.84SYY88 pKa = 11.79NLNPDD93 pKa = 3.97LNHH96 pKa = 7.18DD97 pKa = 3.77EE98 pKa = 4.55CASIFNSVGIYY109 pKa = 9.31MIIDD113 pKa = 3.43VNSPLEE119 pKa = 3.99GGYY122 pKa = 10.05INRR125 pKa = 11.84DD126 pKa = 3.81DD127 pKa = 4.67PSSTYY132 pKa = 10.32NSEE135 pKa = 3.67YY136 pKa = 10.17LKK138 pKa = 10.76RR139 pKa = 11.84VFTVIEE145 pKa = 4.36GFKK148 pKa = 10.53SYY150 pKa = 11.03PNTLGFFAGNEE161 pKa = 4.25IINDD165 pKa = 3.91LATASEE171 pKa = 4.2NPQYY175 pKa = 10.74IRR177 pKa = 11.84AVQRR181 pKa = 11.84DD182 pKa = 3.95MKK184 pKa = 10.97QYY186 pKa = 10.02IAQNADD192 pKa = 3.23RR193 pKa = 11.84EE194 pKa = 4.51IPVGYY199 pKa = 9.75SAADD203 pKa = 3.37VRR205 pKa = 11.84DD206 pKa = 3.71VLEE209 pKa = 4.5DD210 pKa = 3.0TWAYY214 pKa = 8.7LQCDD218 pKa = 3.43NSNNGDD224 pKa = 3.42SDD226 pKa = 3.75SRR228 pKa = 11.84SDD230 pKa = 4.05FFGLNSYY237 pKa = 8.04SWCGGDD243 pKa = 4.1ATFQSSGYY251 pKa = 9.85DD252 pKa = 3.27DD253 pKa = 4.27LVSMFSNTTIPVFFSEE269 pKa = 4.57YY270 pKa = 10.4GCIEE274 pKa = 3.68VTPRR278 pKa = 11.84VFDD281 pKa = 3.77EE282 pKa = 4.2VQALYY287 pKa = 10.02GPKK290 pKa = 8.14MTVLSGGLVYY300 pKa = 10.65EE301 pKa = 4.81WTQEE305 pKa = 3.83DD306 pKa = 3.54NGYY309 pKa = 10.25GLVNTTSDD317 pKa = 3.44GDD319 pKa = 3.83AQLTTDD325 pKa = 4.85FEE327 pKa = 4.72NLSSQYY333 pKa = 11.64AKK335 pKa = 11.12LNTTLLQSQNDD346 pKa = 3.85TATSLEE352 pKa = 4.2APQCDD357 pKa = 3.21ASLISSDD364 pKa = 3.8SFSTTFDD371 pKa = 3.33IPSQPDD377 pKa = 3.44GVADD381 pKa = 5.22LISNGVDD388 pKa = 3.16NAPSGSIVSVTQTSVDD404 pKa = 3.18ATVYY408 pKa = 9.46ATGGGKK414 pKa = 9.4ISGLQINPVEE424 pKa = 4.52GANAPGSGGNGGGSGLSTGAAPSGTSTSSNSGNGGSSSSQTSSGSEE470 pKa = 3.74ASNTSGGSNASGGSGGSSTSTGSGSSDD497 pKa = 3.0TTATSSGSAASSTEE511 pKa = 3.88SEE513 pKa = 4.27GTGFRR518 pKa = 11.84VDD520 pKa = 3.37GKK522 pKa = 8.88MASSAGLAAAIGAAAFALL540 pKa = 4.16
MM1 pKa = 7.76APITNFLASSLALLSSTALAANPIKK26 pKa = 10.73VQEE29 pKa = 4.13QEE31 pKa = 4.6FIDD34 pKa = 3.92TKK36 pKa = 9.1TDD38 pKa = 2.94EE39 pKa = 4.34RR40 pKa = 11.84FVIVGVDD47 pKa = 3.48YY48 pKa = 11.34QPGGSAAVGTGNGDD62 pKa = 3.77PLSNADD68 pKa = 3.26EE69 pKa = 4.41CRR71 pKa = 11.84RR72 pKa = 11.84DD73 pKa = 3.39AALMQNMGVNTIRR86 pKa = 11.84SYY88 pKa = 11.79NLNPDD93 pKa = 3.97LNHH96 pKa = 7.18DD97 pKa = 3.77EE98 pKa = 4.55CASIFNSVGIYY109 pKa = 9.31MIIDD113 pKa = 3.43VNSPLEE119 pKa = 3.99GGYY122 pKa = 10.05INRR125 pKa = 11.84DD126 pKa = 3.81DD127 pKa = 4.67PSSTYY132 pKa = 10.32NSEE135 pKa = 3.67YY136 pKa = 10.17LKK138 pKa = 10.76RR139 pKa = 11.84VFTVIEE145 pKa = 4.36GFKK148 pKa = 10.53SYY150 pKa = 11.03PNTLGFFAGNEE161 pKa = 4.25IINDD165 pKa = 3.91LATASEE171 pKa = 4.2NPQYY175 pKa = 10.74IRR177 pKa = 11.84AVQRR181 pKa = 11.84DD182 pKa = 3.95MKK184 pKa = 10.97QYY186 pKa = 10.02IAQNADD192 pKa = 3.23RR193 pKa = 11.84EE194 pKa = 4.51IPVGYY199 pKa = 9.75SAADD203 pKa = 3.37VRR205 pKa = 11.84DD206 pKa = 3.71VLEE209 pKa = 4.5DD210 pKa = 3.0TWAYY214 pKa = 8.7LQCDD218 pKa = 3.43NSNNGDD224 pKa = 3.42SDD226 pKa = 3.75SRR228 pKa = 11.84SDD230 pKa = 4.05FFGLNSYY237 pKa = 8.04SWCGGDD243 pKa = 4.1ATFQSSGYY251 pKa = 9.85DD252 pKa = 3.27DD253 pKa = 4.27LVSMFSNTTIPVFFSEE269 pKa = 4.57YY270 pKa = 10.4GCIEE274 pKa = 3.68VTPRR278 pKa = 11.84VFDD281 pKa = 3.77EE282 pKa = 4.2VQALYY287 pKa = 10.02GPKK290 pKa = 8.14MTVLSGGLVYY300 pKa = 10.65EE301 pKa = 4.81WTQEE305 pKa = 3.83DD306 pKa = 3.54NGYY309 pKa = 10.25GLVNTTSDD317 pKa = 3.44GDD319 pKa = 3.83AQLTTDD325 pKa = 4.85FEE327 pKa = 4.72NLSSQYY333 pKa = 11.64AKK335 pKa = 11.12LNTTLLQSQNDD346 pKa = 3.85TATSLEE352 pKa = 4.2APQCDD357 pKa = 3.21ASLISSDD364 pKa = 3.8SFSTTFDD371 pKa = 3.33IPSQPDD377 pKa = 3.44GVADD381 pKa = 5.22LISNGVDD388 pKa = 3.16NAPSGSIVSVTQTSVDD404 pKa = 3.18ATVYY408 pKa = 9.46ATGGGKK414 pKa = 9.4ISGLQINPVEE424 pKa = 4.52GANAPGSGGNGGGSGLSTGAAPSGTSTSSNSGNGGSSSSQTSSGSEE470 pKa = 3.74ASNTSGGSNASGGSGGSSTSTGSGSSDD497 pKa = 3.0TTATSSGSAASSTEE511 pKa = 3.88SEE513 pKa = 4.27GTGFRR518 pKa = 11.84VDD520 pKa = 3.37GKK522 pKa = 8.88MASSAGLAAAIGAAAFALL540 pKa = 4.16
Molecular weight: 55.72 kDa
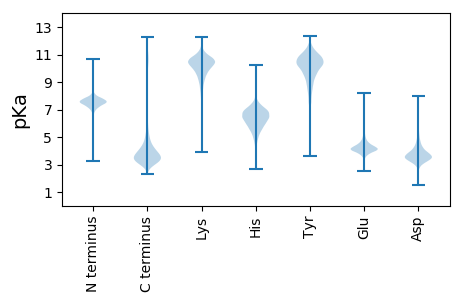
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3M6WIJ7|A0A3M6WIJ7_HORWE Metallophos domain-containing protein OS=Hortaea werneckii OX=91943 GN=D0863_12995 PE=4 SV=1
MM1 pKa = 7.88PSHH4 pKa = 6.89KK5 pKa = 10.44SFRR8 pKa = 11.84TKK10 pKa = 10.65VKK12 pKa = 9.82LAKK15 pKa = 9.79AQKK18 pKa = 8.73QNRR21 pKa = 11.84PIPQWIRR28 pKa = 11.84LRR30 pKa = 11.84TGNTIRR36 pKa = 11.84YY37 pKa = 5.79NAKK40 pKa = 8.89RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 4.15WRR45 pKa = 11.84KK46 pKa = 7.38TRR48 pKa = 11.84IGII51 pKa = 4.0
MM1 pKa = 7.88PSHH4 pKa = 6.89KK5 pKa = 10.44SFRR8 pKa = 11.84TKK10 pKa = 10.65VKK12 pKa = 9.82LAKK15 pKa = 9.79AQKK18 pKa = 8.73QNRR21 pKa = 11.84PIPQWIRR28 pKa = 11.84LRR30 pKa = 11.84TGNTIRR36 pKa = 11.84YY37 pKa = 5.79NAKK40 pKa = 8.89RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 4.15WRR45 pKa = 11.84KK46 pKa = 7.38TRR48 pKa = 11.84IGII51 pKa = 4.0
Molecular weight: 6.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
8038627 |
50 |
4367 |
488.2 |
53.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.126 ± 0.014 | 1.135 ± 0.007 |
5.729 ± 0.014 | 6.432 ± 0.019 |
3.52 ± 0.013 | 7.48 ± 0.017 |
2.384 ± 0.007 | 4.275 ± 0.014 |
4.809 ± 0.019 | 8.42 ± 0.02 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.271 ± 0.007 | 3.63 ± 0.009 |
6.16 ± 0.02 | 4.464 ± 0.015 |
6.065 ± 0.02 | 8.159 ± 0.027 |
5.931 ± 0.015 | 5.845 ± 0.014 |
1.428 ± 0.008 | 2.737 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
