
Devosia sp. Root685
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Devosiaceae; Devosia; unclassified Devosia
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
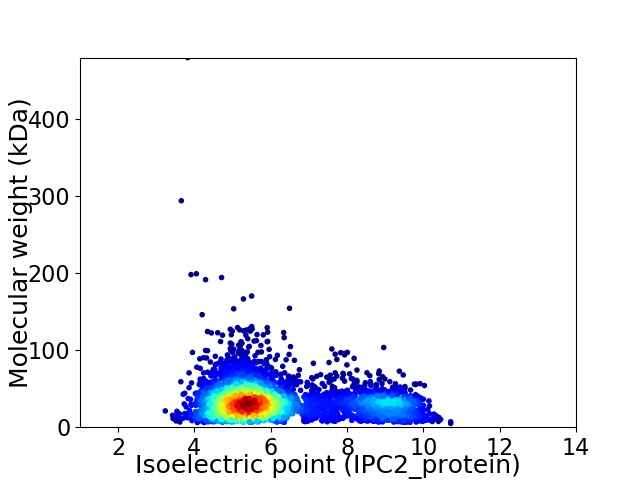
Virtual 2D-PAGE plot for 4002 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q8ICC0|A0A0Q8ICC0_9RHIZ NMT1 domain-containing protein OS=Devosia sp. Root685 OX=1736587 GN=ASD83_20100 PE=4 SV=1
MM1 pKa = 6.74EE2 pKa = 4.39TAATTIAGYY11 pKa = 10.72ALYY14 pKa = 10.54GRR16 pKa = 11.84LEE18 pKa = 4.03GDD20 pKa = 3.32VFYY23 pKa = 10.38FALSSAQPINATSTLWLNTDD43 pKa = 3.36ANINTGYY50 pKa = 10.31KK51 pKa = 9.36VWGFAAGAEE60 pKa = 4.29FNVNFGANGVPRR72 pKa = 11.84LYY74 pKa = 10.4TGADD78 pKa = 3.29GQTLVGDD85 pKa = 4.17LTYY88 pKa = 11.57SMSADD93 pKa = 3.12RR94 pKa = 11.84KK95 pKa = 8.61VLEE98 pKa = 3.97IALPKK103 pKa = 10.31SLLGASVSSVGIMADD118 pKa = 2.97INNTIFLPADD128 pKa = 3.54YY129 pKa = 8.78TQAAYY134 pKa = 9.08TITTPVPSVYY144 pKa = 10.5DD145 pKa = 3.52GLLTEE150 pKa = 4.32WTVEE154 pKa = 3.75QRR156 pKa = 11.84LDD158 pKa = 3.45NTPDD162 pKa = 3.33KK163 pKa = 11.18VVAGYY168 pKa = 9.93EE169 pKa = 4.17LYY171 pKa = 11.12GKK173 pKa = 10.06VADD176 pKa = 4.36DD177 pKa = 3.93SFVFGLKK184 pKa = 10.06SAVAIGPNTTFWLNTDD200 pKa = 3.23NNTATGYY207 pKa = 9.48QIWGFAGGAEE217 pKa = 4.12FNVNIGADD225 pKa = 3.78GVARR229 pKa = 11.84LYY231 pKa = 11.07SGADD235 pKa = 3.21GQTLVGTLEE244 pKa = 3.98YY245 pKa = 10.42KK246 pKa = 10.35IAAGGLSMEE255 pKa = 4.35FAVPKK260 pKa = 10.5ALLGPTVTSVVVLADD275 pKa = 3.28INNSVFLPPSYY286 pKa = 11.04SSGGYY291 pKa = 9.96VLTTPLVVPPGPYY304 pKa = 9.99DD305 pKa = 3.58GLLTEE310 pKa = 4.51WTAAQRR316 pKa = 11.84LDD318 pKa = 3.41SGAGVVAGYY327 pKa = 9.72EE328 pKa = 4.01LYY330 pKa = 9.64GTVSEE335 pKa = 4.81DD336 pKa = 3.14KK337 pKa = 10.84FVFALKK343 pKa = 10.3SAVSIGQNTTFWLNTDD359 pKa = 3.51ANVATGHH366 pKa = 6.04QIFGFAGGAEE376 pKa = 4.11FNVNIGADD384 pKa = 3.78GVARR388 pKa = 11.84LYY390 pKa = 11.07SGADD394 pKa = 3.2GQTLVGEE401 pKa = 4.58IDD403 pKa = 3.92HH404 pKa = 7.05KK405 pKa = 10.66IAPDD409 pKa = 3.51GMSMEE414 pKa = 4.33FAVPRR419 pKa = 11.84SLIGASVTAITLLADD434 pKa = 3.54VNDD437 pKa = 4.28TVFLPTTYY445 pKa = 11.35ANGGYY450 pKa = 10.8ALVDD454 pKa = 3.71PASIPPSQFDD464 pKa = 3.57GVLTEE469 pKa = 4.05WTANQRR475 pKa = 11.84LEE477 pKa = 4.21TPMTSVDD484 pKa = 3.21GYY486 pKa = 10.63EE487 pKa = 4.62FYY489 pKa = 11.16GQYY492 pKa = 10.67SDD494 pKa = 3.92GQFTFGFKK502 pKa = 10.12SAVVIGPNTTFWLNTDD518 pKa = 3.58GNVATGTQVFGYY530 pKa = 10.77AGGAEE535 pKa = 4.14FNVNIGSDD543 pKa = 3.74GVARR547 pKa = 11.84LYY549 pKa = 11.04SGAAGQTLVGAIDD562 pKa = 3.88YY563 pKa = 11.08KK564 pKa = 10.92LGPDD568 pKa = 3.25GKK570 pKa = 8.48TIEE573 pKa = 4.27FAVPKK578 pKa = 10.55AMIGASVTSVSILADD593 pKa = 3.51VNDD596 pKa = 4.5SVFLPANYY604 pKa = 10.03LSPAYY609 pKa = 8.84TVYY612 pKa = 11.03DD613 pKa = 4.08PASLPAVTDD622 pKa = 3.37TGNKK626 pKa = 7.9IAIVFSQTTANNYY639 pKa = 8.38FSQMAYY645 pKa = 9.89SQLVMAAQSQAMAAGIPFDD664 pKa = 4.42LVSEE668 pKa = 4.52ADD670 pKa = 3.81LADD673 pKa = 3.78LSKK676 pKa = 10.4MVNYY680 pKa = 10.04DD681 pKa = 3.76AIVFPSFRR689 pKa = 11.84NVPANYY695 pKa = 8.58TAIYY699 pKa = 10.37DD700 pKa = 3.83VLTKK704 pKa = 10.52LVYY707 pKa = 10.39QYY709 pKa = 11.33DD710 pKa = 3.69VSLIAAGDD718 pKa = 3.8FMTNDD723 pKa = 3.86ANNASLPNNAYY734 pKa = 10.23EE735 pKa = 4.73RR736 pKa = 11.84MQTLFGLNRR745 pKa = 11.84TGGEE749 pKa = 4.03DD750 pKa = 3.46GVTVNIEE757 pKa = 3.82ATPAGHH763 pKa = 7.81AITEE767 pKa = 4.59GYY769 pKa = 8.87GANGAIHH776 pKa = 7.48TYY778 pKa = 9.48TGAATSYY785 pKa = 10.54FSAANPNAGSVTVIAEE801 pKa = 3.95QVVNGTTHH809 pKa = 6.09SAVLGTVTGGRR820 pKa = 11.84NVHH823 pKa = 6.53FSTDD827 pKa = 3.21ALLGDD832 pKa = 4.88LNLLGQAIDD841 pKa = 3.63WVNEE845 pKa = 3.94EE846 pKa = 4.24TGGPTVSLNMTRR858 pKa = 11.84NTSLFASRR866 pKa = 11.84NDD868 pKa = 3.1MDD870 pKa = 4.37QSQEE874 pKa = 4.08TYY876 pKa = 10.84DD877 pKa = 3.77VDD879 pKa = 3.68GGIYY883 pKa = 10.47DD884 pKa = 3.66AMLPFLQQWKK894 pKa = 8.25TDD896 pKa = 3.62YY897 pKa = 11.18NFVGSYY903 pKa = 8.66YY904 pKa = 10.99VNVGFNPPDD913 pKa = 3.39QEE915 pKa = 4.7TNWLISSQYY924 pKa = 8.04YY925 pKa = 10.51DD926 pKa = 3.58AMRR929 pKa = 11.84AMGNEE934 pKa = 4.21IGSHH938 pKa = 6.11SYY940 pKa = 7.77THH942 pKa = 7.36PEE944 pKa = 3.83DD945 pKa = 4.05TNFLLPNVLTQALLDD960 pKa = 3.75QRR962 pKa = 11.84IAQYY966 pKa = 9.02ATRR969 pKa = 11.84PGGPGIIGQSLASMSLADD987 pKa = 4.1INAKK991 pKa = 9.44IAQVLAAPNPSALDD1005 pKa = 3.56ATSKK1009 pKa = 11.14AFLEE1013 pKa = 4.18ATYY1016 pKa = 9.83TFQFATARR1024 pKa = 11.84AVLEE1028 pKa = 4.35ANLGYY1033 pKa = 10.42SIGGAAVPGMPEE1045 pKa = 3.7QLLPAQQIIQYY1056 pKa = 10.01YY1057 pKa = 10.13DD1058 pKa = 3.64YY1059 pKa = 11.15LSGGASMLGAGYY1071 pKa = 9.27PGAIGYY1077 pKa = 7.53LTPGADD1083 pKa = 3.4GQVYY1087 pKa = 10.01IAPNMSFDD1095 pKa = 3.77FTLMGWLGLTVAQAEE1110 pKa = 4.55AKK1112 pKa = 8.64WAAEE1116 pKa = 3.88WSQLTANSDD1125 pKa = 3.46MPIVVWPWHH1134 pKa = 6.95DD1135 pKa = 3.69YY1136 pKa = 11.13GVTSWSLDD1144 pKa = 3.09AGQPSPYY1151 pKa = 8.03TAAMFTNFIAAAYY1164 pKa = 8.88AAGTEE1169 pKa = 4.34FVTLADD1175 pKa = 3.8LAARR1179 pKa = 11.84IKK1181 pKa = 10.79AFEE1184 pKa = 3.99RR1185 pKa = 11.84ADD1187 pKa = 3.62FGFSVAGDD1195 pKa = 4.48VITMHH1200 pKa = 7.05ATPQQGQLGTFALNLDD1216 pKa = 3.95DD1217 pKa = 6.61LGGKK1221 pKa = 7.01TIKK1224 pKa = 10.56SVANWYY1230 pKa = 10.43AYY1232 pKa = 10.39DD1233 pKa = 4.22ADD1235 pKa = 4.88SVFLDD1240 pKa = 3.62ADD1242 pKa = 3.66GGTFTVQLGTTVDD1255 pKa = 4.05DD1256 pKa = 3.93VTHH1259 pKa = 5.94ITSIGSRR1266 pKa = 11.84AQLVSLTGDD1275 pKa = 3.4GTNLDD1280 pKa = 3.59FTIAGEE1286 pKa = 4.36GKK1288 pKa = 8.55VTIDD1292 pKa = 3.36VKK1294 pKa = 9.83QTAGFVYY1301 pKa = 9.9QATGATIVSQTGDD1314 pKa = 3.56ILTLDD1319 pKa = 4.01LGAIGSHH1326 pKa = 4.7TVALRR1331 pKa = 11.84QVAINKK1337 pKa = 9.54APTDD1341 pKa = 3.54IVVSNQVALPEE1352 pKa = 3.93NTAARR1357 pKa = 11.84TKK1359 pKa = 10.4IADD1362 pKa = 4.13LAVIDD1367 pKa = 5.22PDD1369 pKa = 3.8TDD1371 pKa = 3.46PLLRR1375 pKa = 11.84MNVVTLSDD1383 pKa = 3.19ARR1385 pKa = 11.84FEE1387 pKa = 4.39VDD1389 pKa = 3.03ASTGALYY1396 pKa = 10.7LKK1398 pKa = 10.45AGQVVNFEE1406 pKa = 4.34AGATIAMTLTATDD1419 pKa = 4.53GPLSFSKK1426 pKa = 10.9ALVLTVVNVNEE1437 pKa = 4.65APTGAPTVAGAAVEE1451 pKa = 4.13NVLLSSGTALIADD1464 pKa = 4.42PDD1466 pKa = 3.98GLGAFTYY1473 pKa = 9.55QWQRR1477 pKa = 11.84GNGTVFTNIAGATAATYY1494 pKa = 10.85RR1495 pKa = 11.84LVQADD1500 pKa = 3.68ADD1502 pKa = 3.81QLVRR1506 pKa = 11.84VAVSYY1511 pKa = 11.08RR1512 pKa = 11.84DD1513 pKa = 3.48GGATTEE1519 pKa = 4.58TVFSAATTQVVDD1531 pKa = 3.61VLANTTLTPLAANTSRR1547 pKa = 11.84TITTAEE1553 pKa = 3.92LVGTGISGTVTINSLTASIGTLVALGGGQWTYY1585 pKa = 10.83TPPLNNDD1592 pKa = 3.39TAVTFSYY1599 pKa = 9.6TASNGTKK1606 pKa = 10.29LAQGTSAMDD1615 pKa = 4.18LLPSNSIMGTAGVDD1629 pKa = 3.23ILAGRR1634 pKa = 11.84ASADD1638 pKa = 3.72TYY1640 pKa = 11.11HH1641 pKa = 6.9GLAGGDD1647 pKa = 4.0TISGGAGDD1655 pKa = 4.96DD1656 pKa = 3.84IIYY1659 pKa = 10.65GDD1661 pKa = 4.58EE1662 pKa = 4.69GNDD1665 pKa = 3.46TANGGDD1671 pKa = 3.73GNDD1674 pKa = 3.3VFFATLNDD1682 pKa = 3.6GNDD1685 pKa = 3.41RR1686 pKa = 11.84YY1687 pKa = 8.97TGNAGSDD1694 pKa = 3.5RR1695 pKa = 11.84YY1696 pKa = 10.66DD1697 pKa = 3.62LSGTTAAATVNLTNGTATSSQTGSDD1722 pKa = 3.2TLATIEE1728 pKa = 4.32NVTGGSGADD1737 pKa = 3.61TITGSSAANVLNGGAGNDD1755 pKa = 3.28ILLGMGGADD1764 pKa = 3.52TLIGGDD1770 pKa = 3.42GADD1773 pKa = 4.08RR1774 pKa = 11.84LTGGAGRR1781 pKa = 11.84DD1782 pKa = 3.81VMTGGADD1789 pKa = 3.18NDD1791 pKa = 4.21TFIFTSTAEE1800 pKa = 4.07MGTTATTRR1808 pKa = 11.84DD1809 pKa = 3.9VITDD1813 pKa = 3.76FTPGQDD1819 pKa = 3.25KK1820 pKa = 10.96FDD1822 pKa = 4.55FSAIDD1827 pKa = 3.71ANTMLSGNQAFTFLATQGAAFTGVRR1852 pKa = 11.84GQLNWSVQDD1861 pKa = 3.76AAGTANDD1868 pKa = 3.69KK1869 pKa = 10.29TIVMGDD1875 pKa = 3.51LNGDD1879 pKa = 4.39RR1880 pKa = 11.84IADD1883 pKa = 3.47FHH1885 pKa = 8.43VEE1887 pKa = 3.93LTGLIQLGVGDD1898 pKa = 5.52FILL1901 pKa = 5.14
MM1 pKa = 6.74EE2 pKa = 4.39TAATTIAGYY11 pKa = 10.72ALYY14 pKa = 10.54GRR16 pKa = 11.84LEE18 pKa = 4.03GDD20 pKa = 3.32VFYY23 pKa = 10.38FALSSAQPINATSTLWLNTDD43 pKa = 3.36ANINTGYY50 pKa = 10.31KK51 pKa = 9.36VWGFAAGAEE60 pKa = 4.29FNVNFGANGVPRR72 pKa = 11.84LYY74 pKa = 10.4TGADD78 pKa = 3.29GQTLVGDD85 pKa = 4.17LTYY88 pKa = 11.57SMSADD93 pKa = 3.12RR94 pKa = 11.84KK95 pKa = 8.61VLEE98 pKa = 3.97IALPKK103 pKa = 10.31SLLGASVSSVGIMADD118 pKa = 2.97INNTIFLPADD128 pKa = 3.54YY129 pKa = 8.78TQAAYY134 pKa = 9.08TITTPVPSVYY144 pKa = 10.5DD145 pKa = 3.52GLLTEE150 pKa = 4.32WTVEE154 pKa = 3.75QRR156 pKa = 11.84LDD158 pKa = 3.45NTPDD162 pKa = 3.33KK163 pKa = 11.18VVAGYY168 pKa = 9.93EE169 pKa = 4.17LYY171 pKa = 11.12GKK173 pKa = 10.06VADD176 pKa = 4.36DD177 pKa = 3.93SFVFGLKK184 pKa = 10.06SAVAIGPNTTFWLNTDD200 pKa = 3.23NNTATGYY207 pKa = 9.48QIWGFAGGAEE217 pKa = 4.12FNVNIGADD225 pKa = 3.78GVARR229 pKa = 11.84LYY231 pKa = 11.07SGADD235 pKa = 3.21GQTLVGTLEE244 pKa = 3.98YY245 pKa = 10.42KK246 pKa = 10.35IAAGGLSMEE255 pKa = 4.35FAVPKK260 pKa = 10.5ALLGPTVTSVVVLADD275 pKa = 3.28INNSVFLPPSYY286 pKa = 11.04SSGGYY291 pKa = 9.96VLTTPLVVPPGPYY304 pKa = 9.99DD305 pKa = 3.58GLLTEE310 pKa = 4.51WTAAQRR316 pKa = 11.84LDD318 pKa = 3.41SGAGVVAGYY327 pKa = 9.72EE328 pKa = 4.01LYY330 pKa = 9.64GTVSEE335 pKa = 4.81DD336 pKa = 3.14KK337 pKa = 10.84FVFALKK343 pKa = 10.3SAVSIGQNTTFWLNTDD359 pKa = 3.51ANVATGHH366 pKa = 6.04QIFGFAGGAEE376 pKa = 4.11FNVNIGADD384 pKa = 3.78GVARR388 pKa = 11.84LYY390 pKa = 11.07SGADD394 pKa = 3.2GQTLVGEE401 pKa = 4.58IDD403 pKa = 3.92HH404 pKa = 7.05KK405 pKa = 10.66IAPDD409 pKa = 3.51GMSMEE414 pKa = 4.33FAVPRR419 pKa = 11.84SLIGASVTAITLLADD434 pKa = 3.54VNDD437 pKa = 4.28TVFLPTTYY445 pKa = 11.35ANGGYY450 pKa = 10.8ALVDD454 pKa = 3.71PASIPPSQFDD464 pKa = 3.57GVLTEE469 pKa = 4.05WTANQRR475 pKa = 11.84LEE477 pKa = 4.21TPMTSVDD484 pKa = 3.21GYY486 pKa = 10.63EE487 pKa = 4.62FYY489 pKa = 11.16GQYY492 pKa = 10.67SDD494 pKa = 3.92GQFTFGFKK502 pKa = 10.12SAVVIGPNTTFWLNTDD518 pKa = 3.58GNVATGTQVFGYY530 pKa = 10.77AGGAEE535 pKa = 4.14FNVNIGSDD543 pKa = 3.74GVARR547 pKa = 11.84LYY549 pKa = 11.04SGAAGQTLVGAIDD562 pKa = 3.88YY563 pKa = 11.08KK564 pKa = 10.92LGPDD568 pKa = 3.25GKK570 pKa = 8.48TIEE573 pKa = 4.27FAVPKK578 pKa = 10.55AMIGASVTSVSILADD593 pKa = 3.51VNDD596 pKa = 4.5SVFLPANYY604 pKa = 10.03LSPAYY609 pKa = 8.84TVYY612 pKa = 11.03DD613 pKa = 4.08PASLPAVTDD622 pKa = 3.37TGNKK626 pKa = 7.9IAIVFSQTTANNYY639 pKa = 8.38FSQMAYY645 pKa = 9.89SQLVMAAQSQAMAAGIPFDD664 pKa = 4.42LVSEE668 pKa = 4.52ADD670 pKa = 3.81LADD673 pKa = 3.78LSKK676 pKa = 10.4MVNYY680 pKa = 10.04DD681 pKa = 3.76AIVFPSFRR689 pKa = 11.84NVPANYY695 pKa = 8.58TAIYY699 pKa = 10.37DD700 pKa = 3.83VLTKK704 pKa = 10.52LVYY707 pKa = 10.39QYY709 pKa = 11.33DD710 pKa = 3.69VSLIAAGDD718 pKa = 3.8FMTNDD723 pKa = 3.86ANNASLPNNAYY734 pKa = 10.23EE735 pKa = 4.73RR736 pKa = 11.84MQTLFGLNRR745 pKa = 11.84TGGEE749 pKa = 4.03DD750 pKa = 3.46GVTVNIEE757 pKa = 3.82ATPAGHH763 pKa = 7.81AITEE767 pKa = 4.59GYY769 pKa = 8.87GANGAIHH776 pKa = 7.48TYY778 pKa = 9.48TGAATSYY785 pKa = 10.54FSAANPNAGSVTVIAEE801 pKa = 3.95QVVNGTTHH809 pKa = 6.09SAVLGTVTGGRR820 pKa = 11.84NVHH823 pKa = 6.53FSTDD827 pKa = 3.21ALLGDD832 pKa = 4.88LNLLGQAIDD841 pKa = 3.63WVNEE845 pKa = 3.94EE846 pKa = 4.24TGGPTVSLNMTRR858 pKa = 11.84NTSLFASRR866 pKa = 11.84NDD868 pKa = 3.1MDD870 pKa = 4.37QSQEE874 pKa = 4.08TYY876 pKa = 10.84DD877 pKa = 3.77VDD879 pKa = 3.68GGIYY883 pKa = 10.47DD884 pKa = 3.66AMLPFLQQWKK894 pKa = 8.25TDD896 pKa = 3.62YY897 pKa = 11.18NFVGSYY903 pKa = 8.66YY904 pKa = 10.99VNVGFNPPDD913 pKa = 3.39QEE915 pKa = 4.7TNWLISSQYY924 pKa = 8.04YY925 pKa = 10.51DD926 pKa = 3.58AMRR929 pKa = 11.84AMGNEE934 pKa = 4.21IGSHH938 pKa = 6.11SYY940 pKa = 7.77THH942 pKa = 7.36PEE944 pKa = 3.83DD945 pKa = 4.05TNFLLPNVLTQALLDD960 pKa = 3.75QRR962 pKa = 11.84IAQYY966 pKa = 9.02ATRR969 pKa = 11.84PGGPGIIGQSLASMSLADD987 pKa = 4.1INAKK991 pKa = 9.44IAQVLAAPNPSALDD1005 pKa = 3.56ATSKK1009 pKa = 11.14AFLEE1013 pKa = 4.18ATYY1016 pKa = 9.83TFQFATARR1024 pKa = 11.84AVLEE1028 pKa = 4.35ANLGYY1033 pKa = 10.42SIGGAAVPGMPEE1045 pKa = 3.7QLLPAQQIIQYY1056 pKa = 10.01YY1057 pKa = 10.13DD1058 pKa = 3.64YY1059 pKa = 11.15LSGGASMLGAGYY1071 pKa = 9.27PGAIGYY1077 pKa = 7.53LTPGADD1083 pKa = 3.4GQVYY1087 pKa = 10.01IAPNMSFDD1095 pKa = 3.77FTLMGWLGLTVAQAEE1110 pKa = 4.55AKK1112 pKa = 8.64WAAEE1116 pKa = 3.88WSQLTANSDD1125 pKa = 3.46MPIVVWPWHH1134 pKa = 6.95DD1135 pKa = 3.69YY1136 pKa = 11.13GVTSWSLDD1144 pKa = 3.09AGQPSPYY1151 pKa = 8.03TAAMFTNFIAAAYY1164 pKa = 8.88AAGTEE1169 pKa = 4.34FVTLADD1175 pKa = 3.8LAARR1179 pKa = 11.84IKK1181 pKa = 10.79AFEE1184 pKa = 3.99RR1185 pKa = 11.84ADD1187 pKa = 3.62FGFSVAGDD1195 pKa = 4.48VITMHH1200 pKa = 7.05ATPQQGQLGTFALNLDD1216 pKa = 3.95DD1217 pKa = 6.61LGGKK1221 pKa = 7.01TIKK1224 pKa = 10.56SVANWYY1230 pKa = 10.43AYY1232 pKa = 10.39DD1233 pKa = 4.22ADD1235 pKa = 4.88SVFLDD1240 pKa = 3.62ADD1242 pKa = 3.66GGTFTVQLGTTVDD1255 pKa = 4.05DD1256 pKa = 3.93VTHH1259 pKa = 5.94ITSIGSRR1266 pKa = 11.84AQLVSLTGDD1275 pKa = 3.4GTNLDD1280 pKa = 3.59FTIAGEE1286 pKa = 4.36GKK1288 pKa = 8.55VTIDD1292 pKa = 3.36VKK1294 pKa = 9.83QTAGFVYY1301 pKa = 9.9QATGATIVSQTGDD1314 pKa = 3.56ILTLDD1319 pKa = 4.01LGAIGSHH1326 pKa = 4.7TVALRR1331 pKa = 11.84QVAINKK1337 pKa = 9.54APTDD1341 pKa = 3.54IVVSNQVALPEE1352 pKa = 3.93NTAARR1357 pKa = 11.84TKK1359 pKa = 10.4IADD1362 pKa = 4.13LAVIDD1367 pKa = 5.22PDD1369 pKa = 3.8TDD1371 pKa = 3.46PLLRR1375 pKa = 11.84MNVVTLSDD1383 pKa = 3.19ARR1385 pKa = 11.84FEE1387 pKa = 4.39VDD1389 pKa = 3.03ASTGALYY1396 pKa = 10.7LKK1398 pKa = 10.45AGQVVNFEE1406 pKa = 4.34AGATIAMTLTATDD1419 pKa = 4.53GPLSFSKK1426 pKa = 10.9ALVLTVVNVNEE1437 pKa = 4.65APTGAPTVAGAAVEE1451 pKa = 4.13NVLLSSGTALIADD1464 pKa = 4.42PDD1466 pKa = 3.98GLGAFTYY1473 pKa = 9.55QWQRR1477 pKa = 11.84GNGTVFTNIAGATAATYY1494 pKa = 10.85RR1495 pKa = 11.84LVQADD1500 pKa = 3.68ADD1502 pKa = 3.81QLVRR1506 pKa = 11.84VAVSYY1511 pKa = 11.08RR1512 pKa = 11.84DD1513 pKa = 3.48GGATTEE1519 pKa = 4.58TVFSAATTQVVDD1531 pKa = 3.61VLANTTLTPLAANTSRR1547 pKa = 11.84TITTAEE1553 pKa = 3.92LVGTGISGTVTINSLTASIGTLVALGGGQWTYY1585 pKa = 10.83TPPLNNDD1592 pKa = 3.39TAVTFSYY1599 pKa = 9.6TASNGTKK1606 pKa = 10.29LAQGTSAMDD1615 pKa = 4.18LLPSNSIMGTAGVDD1629 pKa = 3.23ILAGRR1634 pKa = 11.84ASADD1638 pKa = 3.72TYY1640 pKa = 11.11HH1641 pKa = 6.9GLAGGDD1647 pKa = 4.0TISGGAGDD1655 pKa = 4.96DD1656 pKa = 3.84IIYY1659 pKa = 10.65GDD1661 pKa = 4.58EE1662 pKa = 4.69GNDD1665 pKa = 3.46TANGGDD1671 pKa = 3.73GNDD1674 pKa = 3.3VFFATLNDD1682 pKa = 3.6GNDD1685 pKa = 3.41RR1686 pKa = 11.84YY1687 pKa = 8.97TGNAGSDD1694 pKa = 3.5RR1695 pKa = 11.84YY1696 pKa = 10.66DD1697 pKa = 3.62LSGTTAAATVNLTNGTATSSQTGSDD1722 pKa = 3.2TLATIEE1728 pKa = 4.32NVTGGSGADD1737 pKa = 3.61TITGSSAANVLNGGAGNDD1755 pKa = 3.28ILLGMGGADD1764 pKa = 3.52TLIGGDD1770 pKa = 3.42GADD1773 pKa = 4.08RR1774 pKa = 11.84LTGGAGRR1781 pKa = 11.84DD1782 pKa = 3.81VMTGGADD1789 pKa = 3.18NDD1791 pKa = 4.21TFIFTSTAEE1800 pKa = 4.07MGTTATTRR1808 pKa = 11.84DD1809 pKa = 3.9VITDD1813 pKa = 3.76FTPGQDD1819 pKa = 3.25KK1820 pKa = 10.96FDD1822 pKa = 4.55FSAIDD1827 pKa = 3.71ANTMLSGNQAFTFLATQGAAFTGVRR1852 pKa = 11.84GQLNWSVQDD1861 pKa = 3.76AAGTANDD1868 pKa = 3.69KK1869 pKa = 10.29TIVMGDD1875 pKa = 3.51LNGDD1879 pKa = 4.39RR1880 pKa = 11.84IADD1883 pKa = 3.47FHH1885 pKa = 8.43VEE1887 pKa = 3.93LTGLIQLGVGDD1898 pKa = 5.52FILL1901 pKa = 5.14
Molecular weight: 197.86 kDa
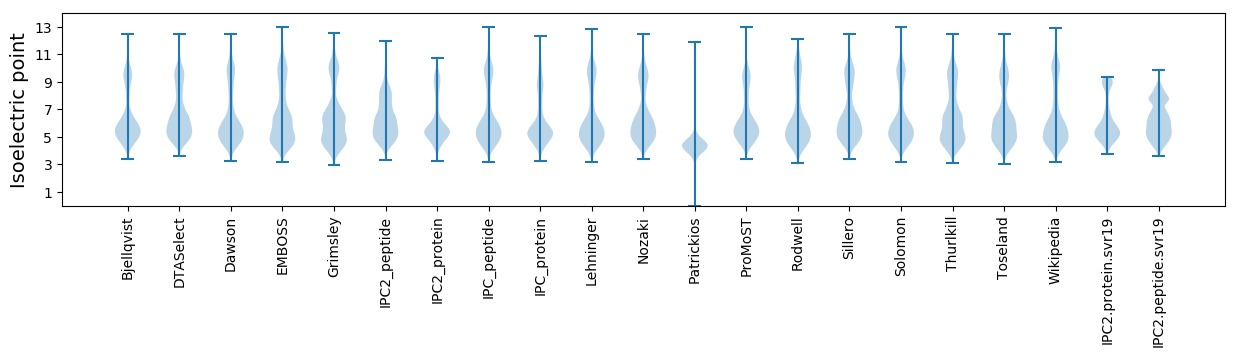
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q8IC93|A0A0Q8IC93_9RHIZ Copper homeostasis protein CutC OS=Devosia sp. Root685 OX=1736587 GN=cutC PE=3 SV=1
MM1 pKa = 7.02VPWRR5 pKa = 11.84KK6 pKa = 7.27TLRR9 pKa = 11.84RR10 pKa = 11.84LDD12 pKa = 3.59GAYY15 pKa = 10.0SEE17 pKa = 4.53HH18 pKa = 6.23TLRR21 pKa = 11.84SYY23 pKa = 11.14RR24 pKa = 11.84SDD26 pKa = 3.09FGAFLRR32 pKa = 11.84WCRR35 pKa = 11.84KK36 pKa = 9.09KK37 pKa = 10.55RR38 pKa = 11.84LKK40 pKa = 10.49ALPAASTTIVAYY52 pKa = 9.78IDD54 pKa = 3.57AEE56 pKa = 4.23GMRR59 pKa = 11.84LKK61 pKa = 10.68PSTVKK66 pKa = 10.35RR67 pKa = 11.84RR68 pKa = 11.84LCALRR73 pKa = 11.84RR74 pKa = 11.84IHH76 pKa = 6.72HH77 pKa = 6.71LSDD80 pKa = 4.47LGDD83 pKa = 3.64PTQSEE88 pKa = 4.76EE89 pKa = 3.83VQLAMRR95 pKa = 11.84RR96 pKa = 11.84VRR98 pKa = 11.84RR99 pKa = 11.84AQPSRR104 pKa = 11.84PKK106 pKa = 9.57QAHH109 pKa = 5.93GVTASLRR116 pKa = 11.84DD117 pKa = 3.43QLLAVCGDD125 pKa = 3.92DD126 pKa = 5.68LIGIRR131 pKa = 11.84DD132 pKa = 3.71KK133 pKa = 11.6VLVSVGFDD141 pKa = 3.19TLCRR145 pKa = 11.84RR146 pKa = 11.84GEE148 pKa = 3.99LVALSIEE155 pKa = 4.51DD156 pKa = 4.41FSPTSDD162 pKa = 2.95GRR164 pKa = 11.84HH165 pKa = 4.09TVLVRR170 pKa = 11.84RR171 pKa = 11.84AKK173 pKa = 10.44NDD175 pKa = 3.41PEE177 pKa = 4.57GAGRR181 pKa = 11.84TAKK184 pKa = 10.27LSKK187 pKa = 10.47ASSDD191 pKa = 4.69VFRR194 pKa = 11.84QWLKK198 pKa = 9.82EE199 pKa = 3.63AGIYY203 pKa = 9.81RR204 pKa = 11.84GPLLRR209 pKa = 11.84PVYY212 pKa = 9.99GNRR215 pKa = 11.84VRR217 pKa = 11.84ALYY220 pKa = 10.44LEE222 pKa = 4.68PLTVGRR228 pKa = 11.84VLKK231 pKa = 10.26KK232 pKa = 10.33LCDD235 pKa = 3.6RR236 pKa = 11.84AGLEE240 pKa = 3.86ALTTEE245 pKa = 4.44RR246 pKa = 11.84VSGHH250 pKa = 5.29SLRR253 pKa = 11.84VGAAQQLTINGFGLPQIMRR272 pKa = 11.84AGGWKK277 pKa = 9.75SVNVVARR284 pKa = 11.84YY285 pKa = 9.44IEE287 pKa = 4.62NVDD290 pKa = 3.59LDD292 pKa = 3.61VWKK295 pKa = 10.81
MM1 pKa = 7.02VPWRR5 pKa = 11.84KK6 pKa = 7.27TLRR9 pKa = 11.84RR10 pKa = 11.84LDD12 pKa = 3.59GAYY15 pKa = 10.0SEE17 pKa = 4.53HH18 pKa = 6.23TLRR21 pKa = 11.84SYY23 pKa = 11.14RR24 pKa = 11.84SDD26 pKa = 3.09FGAFLRR32 pKa = 11.84WCRR35 pKa = 11.84KK36 pKa = 9.09KK37 pKa = 10.55RR38 pKa = 11.84LKK40 pKa = 10.49ALPAASTTIVAYY52 pKa = 9.78IDD54 pKa = 3.57AEE56 pKa = 4.23GMRR59 pKa = 11.84LKK61 pKa = 10.68PSTVKK66 pKa = 10.35RR67 pKa = 11.84RR68 pKa = 11.84LCALRR73 pKa = 11.84RR74 pKa = 11.84IHH76 pKa = 6.72HH77 pKa = 6.71LSDD80 pKa = 4.47LGDD83 pKa = 3.64PTQSEE88 pKa = 4.76EE89 pKa = 3.83VQLAMRR95 pKa = 11.84RR96 pKa = 11.84VRR98 pKa = 11.84RR99 pKa = 11.84AQPSRR104 pKa = 11.84PKK106 pKa = 9.57QAHH109 pKa = 5.93GVTASLRR116 pKa = 11.84DD117 pKa = 3.43QLLAVCGDD125 pKa = 3.92DD126 pKa = 5.68LIGIRR131 pKa = 11.84DD132 pKa = 3.71KK133 pKa = 11.6VLVSVGFDD141 pKa = 3.19TLCRR145 pKa = 11.84RR146 pKa = 11.84GEE148 pKa = 3.99LVALSIEE155 pKa = 4.51DD156 pKa = 4.41FSPTSDD162 pKa = 2.95GRR164 pKa = 11.84HH165 pKa = 4.09TVLVRR170 pKa = 11.84RR171 pKa = 11.84AKK173 pKa = 10.44NDD175 pKa = 3.41PEE177 pKa = 4.57GAGRR181 pKa = 11.84TAKK184 pKa = 10.27LSKK187 pKa = 10.47ASSDD191 pKa = 4.69VFRR194 pKa = 11.84QWLKK198 pKa = 9.82EE199 pKa = 3.63AGIYY203 pKa = 9.81RR204 pKa = 11.84GPLLRR209 pKa = 11.84PVYY212 pKa = 9.99GNRR215 pKa = 11.84VRR217 pKa = 11.84ALYY220 pKa = 10.44LEE222 pKa = 4.68PLTVGRR228 pKa = 11.84VLKK231 pKa = 10.26KK232 pKa = 10.33LCDD235 pKa = 3.6RR236 pKa = 11.84AGLEE240 pKa = 3.86ALTTEE245 pKa = 4.44RR246 pKa = 11.84VSGHH250 pKa = 5.29SLRR253 pKa = 11.84VGAAQQLTINGFGLPQIMRR272 pKa = 11.84AGGWKK277 pKa = 9.75SVNVVARR284 pKa = 11.84YY285 pKa = 9.44IEE287 pKa = 4.62NVDD290 pKa = 3.59LDD292 pKa = 3.61VWKK295 pKa = 10.81
Molecular weight: 33.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1280198 |
41 |
4685 |
319.9 |
34.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.053 ± 0.05 | 0.641 ± 0.01 |
5.698 ± 0.032 | 5.673 ± 0.04 |
3.967 ± 0.024 | 8.609 ± 0.045 |
1.979 ± 0.019 | 5.596 ± 0.027 |
3.268 ± 0.031 | 10.244 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.539 ± 0.019 | 2.915 ± 0.029 |
4.992 ± 0.027 | 3.183 ± 0.02 |
6.279 ± 0.044 | 5.515 ± 0.027 |
5.558 ± 0.035 | 7.638 ± 0.032 |
1.343 ± 0.017 | 2.308 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
