
Mycoplasma cloacale
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Tenericutes; Mollicutes; Mycoplasmatales; Mycoplasmataceae; Mycoplasma
Average proteome isoelectric point is 7.04
Get precalculated fractions of proteins
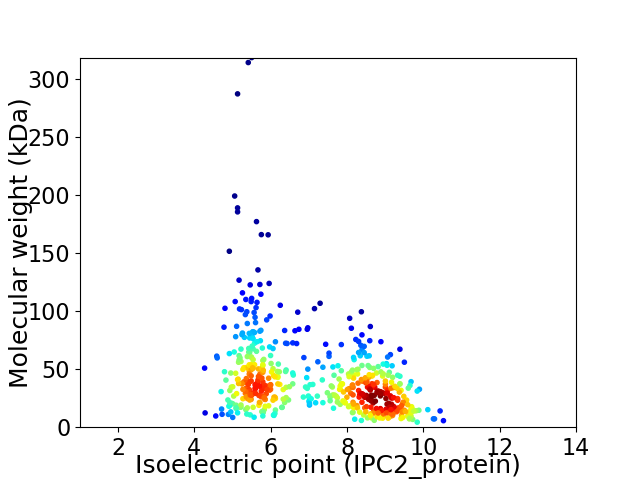
Virtual 2D-PAGE plot for 539 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z4LMB3|A0A2Z4LMB3_9MOLU Uncharacterized protein OS=Mycoplasma cloacale OX=92401 GN=DK849_02485 PE=4 SV=1
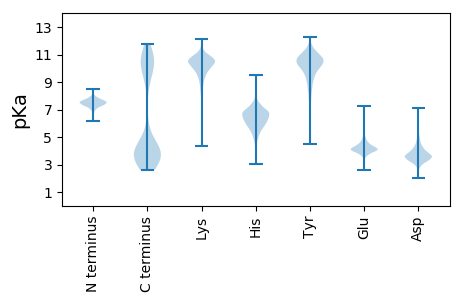
MM1 pKa = 7.34TNKK4 pKa = 10.19KK5 pKa = 10.0YY6 pKa = 10.62FPLTKK11 pKa = 10.37EE12 pKa = 3.95EE13 pKa = 4.25LKK15 pKa = 11.08EE16 pKa = 3.82LVNNLDD22 pKa = 3.84IYY24 pKa = 11.17LGDD27 pKa = 3.69IDD29 pKa = 4.12TSRR32 pKa = 11.84MFDD35 pKa = 3.23MSSLFQNSLRR45 pKa = 11.84KK46 pKa = 9.98KK47 pKa = 10.15FDD49 pKa = 4.72GIEE52 pKa = 3.82TWNTSNVLNMEE63 pKa = 4.2RR64 pKa = 11.84MFEE67 pKa = 3.99SSFINKK73 pKa = 8.73PLNFDD78 pKa = 3.53TSNVKK83 pKa = 10.3NMEE86 pKa = 4.01YY87 pKa = 10.03MFADD91 pKa = 4.62CYY93 pKa = 11.22LFNQPINFNTSKK105 pKa = 10.34VKK107 pKa = 10.56NMKK110 pKa = 10.5NMFSNCYY117 pKa = 9.87FFNQNLNWDD126 pKa = 3.83TSNVKK131 pKa = 10.19NMSYY135 pKa = 9.54MFEE138 pKa = 4.1NCHH141 pKa = 6.75SFNQPLNWDD150 pKa = 3.67TSNVEE155 pKa = 3.63NMSYY159 pKa = 9.64MFRR162 pKa = 11.84GCHH165 pKa = 6.06SFNQPLDD172 pKa = 3.39WDD174 pKa = 4.25TTNLTDD180 pKa = 3.69TYY182 pKa = 11.5SMFSGCRR189 pKa = 11.84SFNQPLNWDD198 pKa = 3.79TTNVEE203 pKa = 3.98DD204 pKa = 3.57MSYY207 pKa = 9.58MFEE210 pKa = 4.21YY211 pKa = 10.36CHH213 pKa = 7.11SFNQPLNWDD222 pKa = 3.67TSNVEE227 pKa = 3.88NMSLMFYY234 pKa = 9.93YY235 pKa = 10.28CHH237 pKa = 7.29SFNQPLNWDD246 pKa = 3.72TSNVEE251 pKa = 4.44DD252 pKa = 3.71MSKK255 pKa = 9.4MFEE258 pKa = 3.94YY259 pKa = 10.26CYY261 pKa = 10.96SFNQPLDD268 pKa = 3.35WDD270 pKa = 3.99TSNVEE275 pKa = 4.28DD276 pKa = 4.1MSDD279 pKa = 3.06MFEE282 pKa = 4.19NCHH285 pKa = 6.73SFNQPLNWDD294 pKa = 3.67TSNVEE299 pKa = 3.77NMAGMFKK306 pKa = 10.37NCHH309 pKa = 6.03SFNQPLDD316 pKa = 3.46WDD318 pKa = 3.93TSNVKK323 pKa = 10.42DD324 pKa = 3.81MNFMFCEE331 pKa = 4.56CSAFNQPLDD340 pKa = 3.39WDD342 pKa = 4.08TTNVEE347 pKa = 3.8NMEE350 pKa = 4.1YY351 pKa = 10.05MFWNCKK357 pKa = 6.9EE358 pKa = 4.56LEE360 pKa = 4.06YY361 pKa = 11.18EE362 pKa = 4.55MNFKK366 pKa = 9.95MPYY369 pKa = 10.12NKK371 pKa = 9.03TKK373 pKa = 11.09NMFHH377 pKa = 6.71GCILLSNEE385 pKa = 4.16EE386 pKa = 4.1EE387 pKa = 4.29DD388 pKa = 5.34NEE390 pKa = 4.56TKK392 pKa = 10.42HH393 pKa = 6.42SKK395 pKa = 9.2EE396 pKa = 3.61WFNDD400 pKa = 3.02QKK402 pKa = 11.17EE403 pKa = 4.1IDD405 pKa = 4.88EE406 pKa = 4.48IPLLKK411 pKa = 10.47FKK413 pKa = 10.58IIEE416 pKa = 4.33NDD418 pKa = 3.83DD419 pKa = 3.71DD420 pKa = 5.12DD421 pKa = 4.88EE422 pKa = 4.35NN423 pKa = 4.41
MM1 pKa = 7.34TNKK4 pKa = 10.19KK5 pKa = 10.0YY6 pKa = 10.62FPLTKK11 pKa = 10.37EE12 pKa = 3.95EE13 pKa = 4.25LKK15 pKa = 11.08EE16 pKa = 3.82LVNNLDD22 pKa = 3.84IYY24 pKa = 11.17LGDD27 pKa = 3.69IDD29 pKa = 4.12TSRR32 pKa = 11.84MFDD35 pKa = 3.23MSSLFQNSLRR45 pKa = 11.84KK46 pKa = 9.98KK47 pKa = 10.15FDD49 pKa = 4.72GIEE52 pKa = 3.82TWNTSNVLNMEE63 pKa = 4.2RR64 pKa = 11.84MFEE67 pKa = 3.99SSFINKK73 pKa = 8.73PLNFDD78 pKa = 3.53TSNVKK83 pKa = 10.3NMEE86 pKa = 4.01YY87 pKa = 10.03MFADD91 pKa = 4.62CYY93 pKa = 11.22LFNQPINFNTSKK105 pKa = 10.34VKK107 pKa = 10.56NMKK110 pKa = 10.5NMFSNCYY117 pKa = 9.87FFNQNLNWDD126 pKa = 3.83TSNVKK131 pKa = 10.19NMSYY135 pKa = 9.54MFEE138 pKa = 4.1NCHH141 pKa = 6.75SFNQPLNWDD150 pKa = 3.67TSNVEE155 pKa = 3.63NMSYY159 pKa = 9.64MFRR162 pKa = 11.84GCHH165 pKa = 6.06SFNQPLDD172 pKa = 3.39WDD174 pKa = 4.25TTNLTDD180 pKa = 3.69TYY182 pKa = 11.5SMFSGCRR189 pKa = 11.84SFNQPLNWDD198 pKa = 3.79TTNVEE203 pKa = 3.98DD204 pKa = 3.57MSYY207 pKa = 9.58MFEE210 pKa = 4.21YY211 pKa = 10.36CHH213 pKa = 7.11SFNQPLNWDD222 pKa = 3.67TSNVEE227 pKa = 3.88NMSLMFYY234 pKa = 9.93YY235 pKa = 10.28CHH237 pKa = 7.29SFNQPLNWDD246 pKa = 3.72TSNVEE251 pKa = 4.44DD252 pKa = 3.71MSKK255 pKa = 9.4MFEE258 pKa = 3.94YY259 pKa = 10.26CYY261 pKa = 10.96SFNQPLDD268 pKa = 3.35WDD270 pKa = 3.99TSNVEE275 pKa = 4.28DD276 pKa = 4.1MSDD279 pKa = 3.06MFEE282 pKa = 4.19NCHH285 pKa = 6.73SFNQPLNWDD294 pKa = 3.67TSNVEE299 pKa = 3.77NMAGMFKK306 pKa = 10.37NCHH309 pKa = 6.03SFNQPLDD316 pKa = 3.46WDD318 pKa = 3.93TSNVKK323 pKa = 10.42DD324 pKa = 3.81MNFMFCEE331 pKa = 4.56CSAFNQPLDD340 pKa = 3.39WDD342 pKa = 4.08TTNVEE347 pKa = 3.8NMEE350 pKa = 4.1YY351 pKa = 10.05MFWNCKK357 pKa = 6.9EE358 pKa = 4.56LEE360 pKa = 4.06YY361 pKa = 11.18EE362 pKa = 4.55MNFKK366 pKa = 9.95MPYY369 pKa = 10.12NKK371 pKa = 9.03TKK373 pKa = 11.09NMFHH377 pKa = 6.71GCILLSNEE385 pKa = 4.16EE386 pKa = 4.1EE387 pKa = 4.29DD388 pKa = 5.34NEE390 pKa = 4.56TKK392 pKa = 10.42HH393 pKa = 6.42SKK395 pKa = 9.2EE396 pKa = 3.61WFNDD400 pKa = 3.02QKK402 pKa = 11.17EE403 pKa = 4.1IDD405 pKa = 4.88EE406 pKa = 4.48IPLLKK411 pKa = 10.47FKK413 pKa = 10.58IIEE416 pKa = 4.33NDD418 pKa = 3.83DD419 pKa = 3.71DD420 pKa = 5.12DD421 pKa = 4.88EE422 pKa = 4.35NN423 pKa = 4.41
Molecular weight: 50.86 kDa
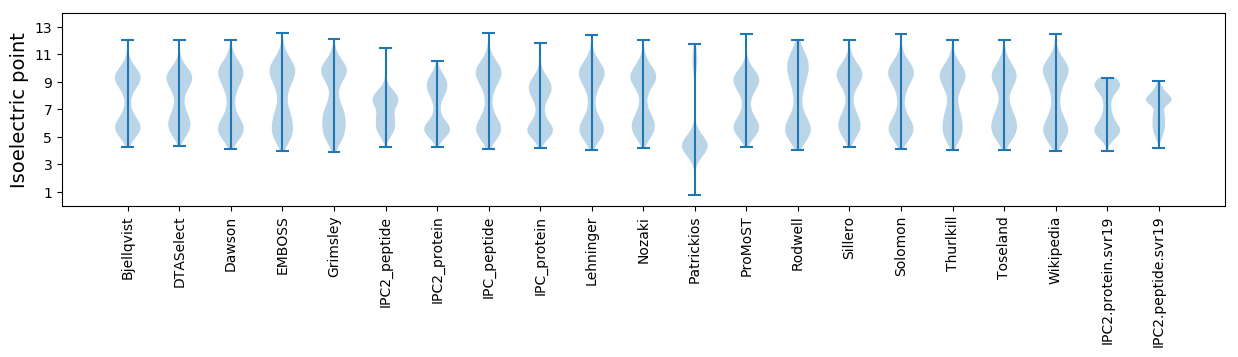
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z4LLZ1|A0A2Z4LLZ1_9MOLU 30S ribosomal protein S5 OS=Mycoplasma cloacale OX=92401 GN=rpsE PE=3 SV=1
MM1 pKa = 7.33ARR3 pKa = 11.84KK4 pKa = 9.63ALMVKK9 pKa = 10.38AEE11 pKa = 4.31RR12 pKa = 11.84EE13 pKa = 4.05PKK15 pKa = 10.35FGVRR19 pKa = 11.84KK20 pKa = 7.54YY21 pKa = 8.6TRR23 pKa = 11.84CQICGRR29 pKa = 11.84VHH31 pKa = 6.76AVLRR35 pKa = 11.84KK36 pKa = 9.58YY37 pKa = 10.23KK38 pKa = 9.85ICRR41 pKa = 11.84ICFRR45 pKa = 11.84EE46 pKa = 3.88LAHH49 pKa = 6.54EE50 pKa = 4.41GKK52 pKa = 10.21IPGVKK57 pKa = 9.38KK58 pKa = 10.89ASWW61 pKa = 3.03
MM1 pKa = 7.33ARR3 pKa = 11.84KK4 pKa = 9.63ALMVKK9 pKa = 10.38AEE11 pKa = 4.31RR12 pKa = 11.84EE13 pKa = 4.05PKK15 pKa = 10.35FGVRR19 pKa = 11.84KK20 pKa = 7.54YY21 pKa = 8.6TRR23 pKa = 11.84CQICGRR29 pKa = 11.84VHH31 pKa = 6.76AVLRR35 pKa = 11.84KK36 pKa = 9.58YY37 pKa = 10.23KK38 pKa = 9.85ICRR41 pKa = 11.84ICFRR45 pKa = 11.84EE46 pKa = 3.88LAHH49 pKa = 6.54EE50 pKa = 4.41GKK52 pKa = 10.21IPGVKK57 pKa = 9.38KK58 pKa = 10.89ASWW61 pKa = 3.03
Molecular weight: 7.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
201187 |
37 |
2772 |
373.3 |
42.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.257 ± 0.116 | 0.596 ± 0.03 |
5.439 ± 0.082 | 7.068 ± 0.106 |
5.055 ± 0.107 | 4.324 ± 0.105 |
1.614 ± 0.037 | 10.169 ± 0.124 |
9.773 ± 0.092 | 9.598 ± 0.092 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.882 ± 0.051 | 8.237 ± 0.166 |
2.656 ± 0.058 | 3.232 ± 0.049 |
2.889 ± 0.059 | 6.26 ± 0.075 |
5.301 ± 0.072 | 5.244 ± 0.071 |
0.941 ± 0.036 | 4.463 ± 0.072 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
