
Human mastadenovirus C
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Preplasmiviricota; Tectiliviricetes; Rowavirales; Adenoviridae; Mastadenovirus
Average proteome isoelectric point is 6.81
Get precalculated fractions of proteins
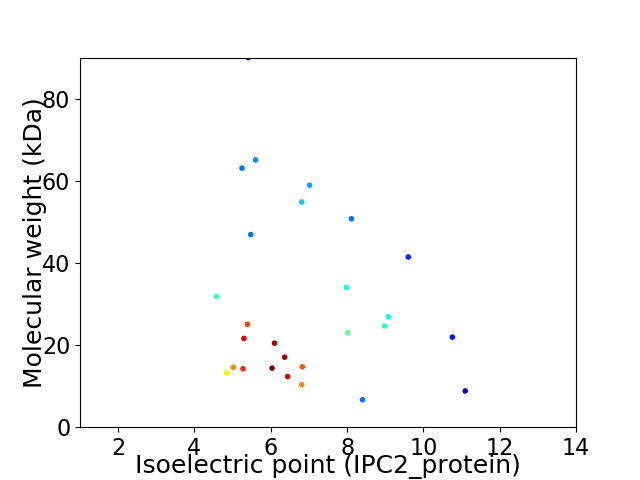
Virtual 2D-PAGE plot for 27 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
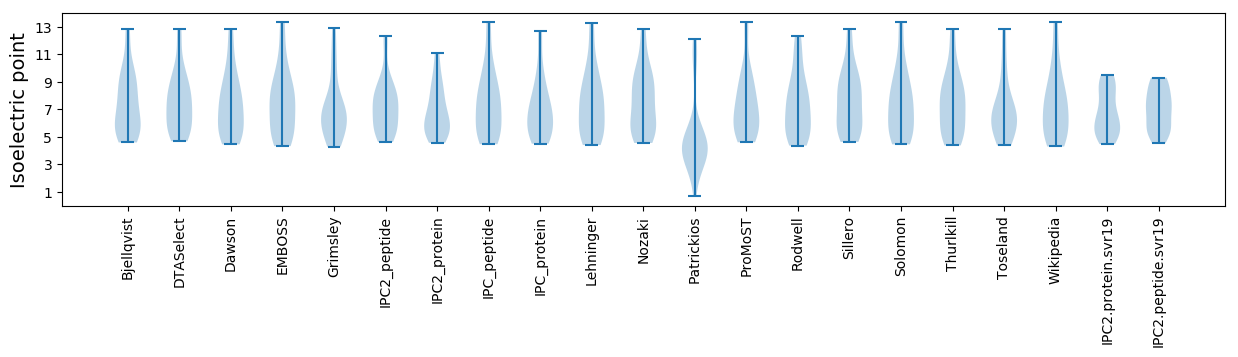
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3Q9HL43|A0A3Q9HL43_9ADEN 33K OS=Human mastadenovirus C OX=129951 PE=3 SV=1
MM1 pKa = 7.5RR2 pKa = 11.84HH3 pKa = 6.51IICHH7 pKa = 5.93GGVITEE13 pKa = 4.22EE14 pKa = 4.15MAASLLDD21 pKa = 3.6QLIEE25 pKa = 4.19EE26 pKa = 4.48VLADD30 pKa = 3.85NLPPPSHH37 pKa = 7.16FEE39 pKa = 3.8PPTLHH44 pKa = 6.78EE45 pKa = 5.5LYY47 pKa = 10.58DD48 pKa = 4.45LDD50 pKa = 3.62VTAPEE55 pKa = 4.89DD56 pKa = 3.83PNEE59 pKa = 4.13EE60 pKa = 3.85AVSQIFPEE68 pKa = 4.34SVMLAVQEE76 pKa = 4.97GIDD79 pKa = 4.38LFTFPPAPGSPEE91 pKa = 3.95PPHH94 pKa = 7.1LSRR97 pKa = 11.84QPEE100 pKa = 4.1QPEE103 pKa = 3.86QRR105 pKa = 11.84ALGPVSMPNLVPEE118 pKa = 4.78VIDD121 pKa = 3.89LTCHH125 pKa = 5.71EE126 pKa = 5.16AGFPPSDD133 pKa = 4.22DD134 pKa = 3.37EE135 pKa = 5.17DD136 pKa = 4.38EE137 pKa = 4.52EE138 pKa = 5.43GEE140 pKa = 4.43EE141 pKa = 4.08FVLDD145 pKa = 4.04YY146 pKa = 11.29VEE148 pKa = 5.48HH149 pKa = 7.23PGHH152 pKa = 6.6GCRR155 pKa = 11.84SCHH158 pKa = 4.33YY159 pKa = 9.82HH160 pKa = 6.49RR161 pKa = 11.84RR162 pKa = 11.84NTGDD166 pKa = 3.82PDD168 pKa = 3.74IMCSLCYY175 pKa = 9.84MRR177 pKa = 11.84TCGMFVYY184 pKa = 10.41SPVSEE189 pKa = 4.7PEE191 pKa = 4.08PEE193 pKa = 4.37PEE195 pKa = 4.57PEE197 pKa = 4.28PEE199 pKa = 3.77PARR202 pKa = 11.84PARR205 pKa = 11.84RR206 pKa = 11.84PKK208 pKa = 9.45MVPAILRR215 pKa = 11.84RR216 pKa = 11.84PTSPVSRR223 pKa = 11.84EE224 pKa = 4.32CNSSTDD230 pKa = 3.44SCDD233 pKa = 3.3SGPSNTPPEE242 pKa = 4.07IHH244 pKa = 6.71PVVPLCPIKK253 pKa = 10.4PVAVRR258 pKa = 11.84VGGRR262 pKa = 11.84RR263 pKa = 11.84RR264 pKa = 11.84AVEE267 pKa = 4.43CIEE270 pKa = 5.02DD271 pKa = 4.03LLNEE275 pKa = 4.64PGQPLDD281 pKa = 4.25LSCKK285 pKa = 10.03RR286 pKa = 11.84PRR288 pKa = 11.84PP289 pKa = 3.69
MM1 pKa = 7.5RR2 pKa = 11.84HH3 pKa = 6.51IICHH7 pKa = 5.93GGVITEE13 pKa = 4.22EE14 pKa = 4.15MAASLLDD21 pKa = 3.6QLIEE25 pKa = 4.19EE26 pKa = 4.48VLADD30 pKa = 3.85NLPPPSHH37 pKa = 7.16FEE39 pKa = 3.8PPTLHH44 pKa = 6.78EE45 pKa = 5.5LYY47 pKa = 10.58DD48 pKa = 4.45LDD50 pKa = 3.62VTAPEE55 pKa = 4.89DD56 pKa = 3.83PNEE59 pKa = 4.13EE60 pKa = 3.85AVSQIFPEE68 pKa = 4.34SVMLAVQEE76 pKa = 4.97GIDD79 pKa = 4.38LFTFPPAPGSPEE91 pKa = 3.95PPHH94 pKa = 7.1LSRR97 pKa = 11.84QPEE100 pKa = 4.1QPEE103 pKa = 3.86QRR105 pKa = 11.84ALGPVSMPNLVPEE118 pKa = 4.78VIDD121 pKa = 3.89LTCHH125 pKa = 5.71EE126 pKa = 5.16AGFPPSDD133 pKa = 4.22DD134 pKa = 3.37EE135 pKa = 5.17DD136 pKa = 4.38EE137 pKa = 4.52EE138 pKa = 5.43GEE140 pKa = 4.43EE141 pKa = 4.08FVLDD145 pKa = 4.04YY146 pKa = 11.29VEE148 pKa = 5.48HH149 pKa = 7.23PGHH152 pKa = 6.6GCRR155 pKa = 11.84SCHH158 pKa = 4.33YY159 pKa = 9.82HH160 pKa = 6.49RR161 pKa = 11.84RR162 pKa = 11.84NTGDD166 pKa = 3.82PDD168 pKa = 3.74IMCSLCYY175 pKa = 9.84MRR177 pKa = 11.84TCGMFVYY184 pKa = 10.41SPVSEE189 pKa = 4.7PEE191 pKa = 4.08PEE193 pKa = 4.37PEE195 pKa = 4.57PEE197 pKa = 4.28PEE199 pKa = 3.77PARR202 pKa = 11.84PARR205 pKa = 11.84RR206 pKa = 11.84PKK208 pKa = 9.45MVPAILRR215 pKa = 11.84RR216 pKa = 11.84PTSPVSRR223 pKa = 11.84EE224 pKa = 4.32CNSSTDD230 pKa = 3.44SCDD233 pKa = 3.3SGPSNTPPEE242 pKa = 4.07IHH244 pKa = 6.71PVVPLCPIKK253 pKa = 10.4PVAVRR258 pKa = 11.84VGGRR262 pKa = 11.84RR263 pKa = 11.84RR264 pKa = 11.84AVEE267 pKa = 4.43CIEE270 pKa = 5.02DD271 pKa = 4.03LLNEE275 pKa = 4.64PGQPLDD281 pKa = 4.25LSCKK285 pKa = 10.03RR286 pKa = 11.84PRR288 pKa = 11.84PP289 pKa = 3.69
Molecular weight: 31.93 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E1ARR7|E1ARR7_9ADEN Isoform of A0A3Q9HL75 Pre-hexon-linking protein VIII OS=Human mastadenovirus C OX=129951 GN=L4 PE=2 SV=1
MM1 pKa = 7.77ALTCRR6 pKa = 11.84LRR8 pKa = 11.84FPVPGFRR15 pKa = 11.84GRR17 pKa = 11.84MHH19 pKa = 7.55RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84GMAGHH27 pKa = 7.2GLTGGMRR34 pKa = 11.84RR35 pKa = 11.84AHH37 pKa = 5.79HH38 pKa = 5.82RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84ASHH45 pKa = 5.58RR46 pKa = 11.84RR47 pKa = 11.84MRR49 pKa = 11.84GGILPLLIPLIAAAIGAVPGIASVALQAQRR79 pKa = 11.84HH80 pKa = 4.41
MM1 pKa = 7.77ALTCRR6 pKa = 11.84LRR8 pKa = 11.84FPVPGFRR15 pKa = 11.84GRR17 pKa = 11.84MHH19 pKa = 7.55RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84GMAGHH27 pKa = 7.2GLTGGMRR34 pKa = 11.84RR35 pKa = 11.84AHH37 pKa = 5.79HH38 pKa = 5.82RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84ASHH45 pKa = 5.58RR46 pKa = 11.84RR47 pKa = 11.84MRR49 pKa = 11.84GGILPLLIPLIAAAIGAVPGIASVALQAQRR79 pKa = 11.84HH80 pKa = 4.41
Molecular weight: 8.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
7412 |
61 |
807 |
274.5 |
30.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.338 ± 0.448 | 2.024 ± 0.403 |
4.79 ± 0.291 | 6.827 ± 0.473 |
3.319 ± 0.222 | 6.152 ± 0.307 |
2.59 ± 0.234 | 3.791 ± 0.281 |
3.386 ± 0.368 | 9.458 ± 0.47 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.604 ± 0.231 | 3.859 ± 0.378 |
7.366 ± 0.486 | 4.439 ± 0.327 |
8.5 ± 0.531 | 6.935 ± 0.372 |
5.491 ± 0.428 | 6.462 ± 0.32 |
1.079 ± 0.133 | 2.59 ± 0.226 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
