
Lake Sarah-associated circular virus-38
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.26
Get precalculated fractions of proteins
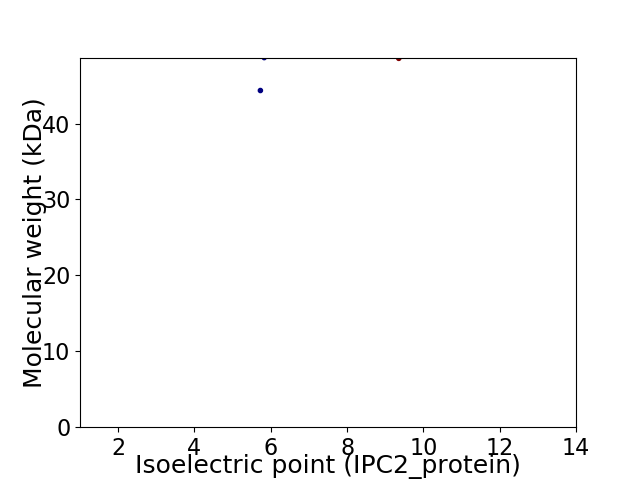
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140AQP9|A0A140AQP9_9VIRU ATP-dependent helicase Rep OS=Lake Sarah-associated circular virus-38 OX=1685766 PE=3 SV=1
MM1 pKa = 7.09SNKK4 pKa = 8.41MAKK7 pKa = 8.4NWCMTLNNYY16 pKa = 9.89DD17 pKa = 3.76DD18 pKa = 5.03SEE20 pKa = 4.36VARR23 pKa = 11.84FEE25 pKa = 4.05SHH27 pKa = 4.75MRR29 pKa = 11.84PWCVYY34 pKa = 8.89YY35 pKa = 9.96IYY37 pKa = 10.84GFEE40 pKa = 4.12RR41 pKa = 11.84GEE43 pKa = 3.92NDD45 pKa = 3.73TPHH48 pKa = 6.33LQCFFSLKK56 pKa = 9.77AKK58 pKa = 10.43KK59 pKa = 10.17RR60 pKa = 11.84MSCLKK65 pKa = 10.39KK66 pKa = 9.94IFPRR70 pKa = 11.84AHH72 pKa = 6.2FEE74 pKa = 4.24VKK76 pKa = 10.16SRR78 pKa = 11.84ASTMEE83 pKa = 3.77QASDD87 pKa = 3.67YY88 pKa = 10.66CKK90 pKa = 10.57KK91 pKa = 10.56EE92 pKa = 3.84EE93 pKa = 4.8NFIEE97 pKa = 4.39WGVLPDD103 pKa = 3.41NSTARR108 pKa = 11.84GLKK111 pKa = 10.2AISDD115 pKa = 4.0NYY117 pKa = 10.69EE118 pKa = 3.86EE119 pKa = 4.33TVEE122 pKa = 4.0LAKK125 pKa = 10.95KK126 pKa = 10.71GDD128 pKa = 3.47IEE130 pKa = 4.98AINPEE135 pKa = 4.48HH136 pKa = 6.14VLKK139 pKa = 10.78YY140 pKa = 10.1YY141 pKa = 8.54PTIKK145 pKa = 10.37RR146 pKa = 11.84IAHH149 pKa = 6.65DD150 pKa = 4.0NKK152 pKa = 10.79KK153 pKa = 9.54MPSDD157 pKa = 3.86LLWEE161 pKa = 4.67EE162 pKa = 4.13GHH164 pKa = 7.07PPNIWIYY171 pKa = 10.79GPTGTGKK178 pKa = 10.22SYY180 pKa = 10.43RR181 pKa = 11.84ARR183 pKa = 11.84AILQEE188 pKa = 4.08NFGQFYY194 pKa = 11.26SKK196 pKa = 10.13MAQNKK201 pKa = 7.55WWDD204 pKa = 3.62KK205 pKa = 10.01YY206 pKa = 11.18DD207 pKa = 3.7GEE209 pKa = 4.37EE210 pKa = 4.24GVLIEE215 pKa = 6.28DD216 pKa = 3.65MDD218 pKa = 3.9ILHH221 pKa = 6.86NYY223 pKa = 6.96MGPYY227 pKa = 8.25MKK229 pKa = 9.78IWADD233 pKa = 3.14KK234 pKa = 9.03YY235 pKa = 10.74AFPVEE240 pKa = 4.52VKK242 pKa = 10.07TSGDD246 pKa = 3.55RR247 pKa = 11.84IRR249 pKa = 11.84PKK251 pKa = 10.78VIVVTSNYY259 pKa = 9.38TIEE262 pKa = 4.58QIWPDD267 pKa = 3.71RR268 pKa = 11.84STHH271 pKa = 5.32GPISRR276 pKa = 11.84RR277 pKa = 11.84FKK279 pKa = 10.35VIHH282 pKa = 6.68MDD284 pKa = 3.6QPWNANINQVLRR296 pKa = 11.84DD297 pKa = 3.95APAEE301 pKa = 3.95TSAPRR306 pKa = 11.84EE307 pKa = 4.2KK308 pKa = 10.21KK309 pKa = 10.38RR310 pKa = 11.84KK311 pKa = 9.08FDD313 pKa = 3.68QPLKK317 pKa = 10.75KK318 pKa = 9.76PALLRR323 pKa = 11.84RR324 pKa = 11.84NAVGDD329 pKa = 3.56LVEE332 pKa = 4.11THH334 pKa = 7.28GYY336 pKa = 9.59QPQQTIPQYY345 pKa = 11.02LEE347 pKa = 3.72PTLEE351 pKa = 3.99EE352 pKa = 3.95LAEE355 pKa = 3.94EE356 pKa = 4.22MEE358 pKa = 4.87IYY360 pKa = 10.2RR361 pKa = 11.84ASQDD365 pKa = 3.22MFEE368 pKa = 4.77EE369 pKa = 4.57LSEE372 pKa = 4.88SDD374 pKa = 5.51DD375 pKa = 5.05LLDD378 pKa = 4.29LL379 pKa = 4.89
MM1 pKa = 7.09SNKK4 pKa = 8.41MAKK7 pKa = 8.4NWCMTLNNYY16 pKa = 9.89DD17 pKa = 3.76DD18 pKa = 5.03SEE20 pKa = 4.36VARR23 pKa = 11.84FEE25 pKa = 4.05SHH27 pKa = 4.75MRR29 pKa = 11.84PWCVYY34 pKa = 8.89YY35 pKa = 9.96IYY37 pKa = 10.84GFEE40 pKa = 4.12RR41 pKa = 11.84GEE43 pKa = 3.92NDD45 pKa = 3.73TPHH48 pKa = 6.33LQCFFSLKK56 pKa = 9.77AKK58 pKa = 10.43KK59 pKa = 10.17RR60 pKa = 11.84MSCLKK65 pKa = 10.39KK66 pKa = 9.94IFPRR70 pKa = 11.84AHH72 pKa = 6.2FEE74 pKa = 4.24VKK76 pKa = 10.16SRR78 pKa = 11.84ASTMEE83 pKa = 3.77QASDD87 pKa = 3.67YY88 pKa = 10.66CKK90 pKa = 10.57KK91 pKa = 10.56EE92 pKa = 3.84EE93 pKa = 4.8NFIEE97 pKa = 4.39WGVLPDD103 pKa = 3.41NSTARR108 pKa = 11.84GLKK111 pKa = 10.2AISDD115 pKa = 4.0NYY117 pKa = 10.69EE118 pKa = 3.86EE119 pKa = 4.33TVEE122 pKa = 4.0LAKK125 pKa = 10.95KK126 pKa = 10.71GDD128 pKa = 3.47IEE130 pKa = 4.98AINPEE135 pKa = 4.48HH136 pKa = 6.14VLKK139 pKa = 10.78YY140 pKa = 10.1YY141 pKa = 8.54PTIKK145 pKa = 10.37RR146 pKa = 11.84IAHH149 pKa = 6.65DD150 pKa = 4.0NKK152 pKa = 10.79KK153 pKa = 9.54MPSDD157 pKa = 3.86LLWEE161 pKa = 4.67EE162 pKa = 4.13GHH164 pKa = 7.07PPNIWIYY171 pKa = 10.79GPTGTGKK178 pKa = 10.22SYY180 pKa = 10.43RR181 pKa = 11.84ARR183 pKa = 11.84AILQEE188 pKa = 4.08NFGQFYY194 pKa = 11.26SKK196 pKa = 10.13MAQNKK201 pKa = 7.55WWDD204 pKa = 3.62KK205 pKa = 10.01YY206 pKa = 11.18DD207 pKa = 3.7GEE209 pKa = 4.37EE210 pKa = 4.24GVLIEE215 pKa = 6.28DD216 pKa = 3.65MDD218 pKa = 3.9ILHH221 pKa = 6.86NYY223 pKa = 6.96MGPYY227 pKa = 8.25MKK229 pKa = 9.78IWADD233 pKa = 3.14KK234 pKa = 9.03YY235 pKa = 10.74AFPVEE240 pKa = 4.52VKK242 pKa = 10.07TSGDD246 pKa = 3.55RR247 pKa = 11.84IRR249 pKa = 11.84PKK251 pKa = 10.78VIVVTSNYY259 pKa = 9.38TIEE262 pKa = 4.58QIWPDD267 pKa = 3.71RR268 pKa = 11.84STHH271 pKa = 5.32GPISRR276 pKa = 11.84RR277 pKa = 11.84FKK279 pKa = 10.35VIHH282 pKa = 6.68MDD284 pKa = 3.6QPWNANINQVLRR296 pKa = 11.84DD297 pKa = 3.95APAEE301 pKa = 3.95TSAPRR306 pKa = 11.84EE307 pKa = 4.2KK308 pKa = 10.21KK309 pKa = 10.38RR310 pKa = 11.84KK311 pKa = 9.08FDD313 pKa = 3.68QPLKK317 pKa = 10.75KK318 pKa = 9.76PALLRR323 pKa = 11.84RR324 pKa = 11.84NAVGDD329 pKa = 3.56LVEE332 pKa = 4.11THH334 pKa = 7.28GYY336 pKa = 9.59QPQQTIPQYY345 pKa = 11.02LEE347 pKa = 3.72PTLEE351 pKa = 3.99EE352 pKa = 3.95LAEE355 pKa = 3.94EE356 pKa = 4.22MEE358 pKa = 4.87IYY360 pKa = 10.2RR361 pKa = 11.84ASQDD365 pKa = 3.22MFEE368 pKa = 4.77EE369 pKa = 4.57LSEE372 pKa = 4.88SDD374 pKa = 5.51DD375 pKa = 5.05LLDD378 pKa = 4.29LL379 pKa = 4.89
Molecular weight: 44.36 kDa
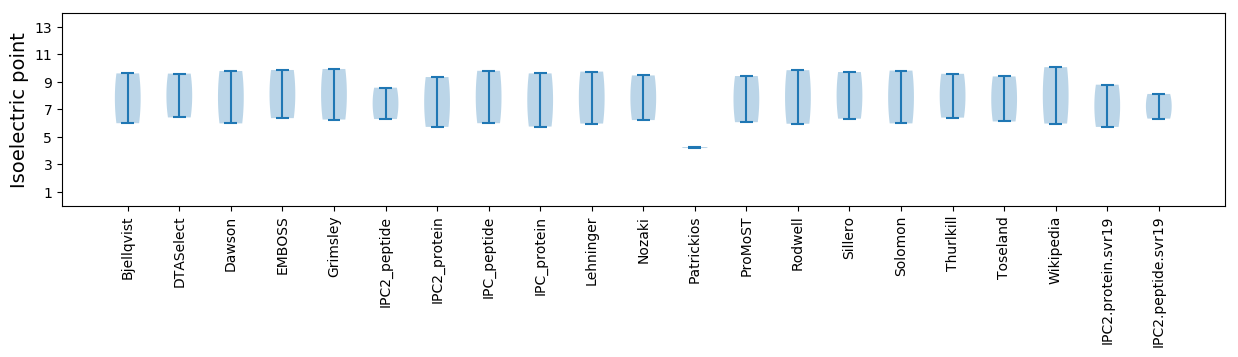
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140AQP9|A0A140AQP9_9VIRU ATP-dependent helicase Rep OS=Lake Sarah-associated circular virus-38 OX=1685766 PE=3 SV=1
MM1 pKa = 8.12DD2 pKa = 5.06GNGDD6 pKa = 3.83VPEE9 pKa = 4.67GWTDD13 pKa = 5.12PITMAEE19 pKa = 3.4IRR21 pKa = 11.84KK22 pKa = 9.32YY23 pKa = 10.47IKK25 pKa = 10.76NSDD28 pKa = 3.48YY29 pKa = 11.64DD30 pKa = 3.56KK31 pKa = 10.92TKK33 pKa = 10.84ARR35 pKa = 11.84AIRR38 pKa = 11.84SLVSRR43 pKa = 11.84RR44 pKa = 11.84PRR46 pKa = 11.84RR47 pKa = 11.84SYY49 pKa = 10.38RR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84SNRR56 pKa = 11.84RR57 pKa = 11.84GSRR60 pKa = 11.84RR61 pKa = 11.84RR62 pKa = 11.84GYY64 pKa = 9.91RR65 pKa = 11.84GYY67 pKa = 10.61GAYY70 pKa = 9.85SRR72 pKa = 11.84RR73 pKa = 11.84GSASGSIGQRR83 pKa = 11.84LGGYY87 pKa = 9.47LGTMLGGAAQNLLKK101 pKa = 10.12TYY103 pKa = 9.06TGLGAYY109 pKa = 7.24QVKK112 pKa = 10.3GNALMPGAVISNPNPHH128 pKa = 6.69GGQVFRR134 pKa = 11.84GSDD137 pKa = 3.48YY138 pKa = 11.33LGDD141 pKa = 3.65IFSSPVAGQFTNQSFPINAALEE163 pKa = 4.05QTFPKK168 pKa = 10.31LAQFLQNFDD177 pKa = 3.39QYY179 pKa = 11.43VIEE182 pKa = 4.09GLIFEE187 pKa = 4.77FRR189 pKa = 11.84SMSCDD194 pKa = 3.11SLNSTNTALGSIIAACNYY212 pKa = 10.03NVLQAPFASKK222 pKa = 10.71AAMEE226 pKa = 4.3EE227 pKa = 4.46YY228 pKa = 10.45EE229 pKa = 4.59GGVSQRR235 pKa = 11.84PSSNMQFFVEE245 pKa = 4.64CARR248 pKa = 11.84AQSPMDD254 pKa = 3.39VLYY257 pKa = 10.0TRR259 pKa = 11.84TGAIPANADD268 pKa = 2.9LRR270 pKa = 11.84MYY272 pKa = 11.05DD273 pKa = 3.88LGNFQIASQGLQGTSVNMGEE293 pKa = 4.37LWVHH297 pKa = 5.07YY298 pKa = 9.65QVAGLKK304 pKa = 9.35EE305 pKa = 3.95KK306 pKa = 10.3IYY308 pKa = 11.07AGFGNYY314 pKa = 8.69NSAYY318 pKa = 9.4RR319 pKa = 11.84AVGSGGNNSNPLGNTRR335 pKa = 11.84TDD337 pKa = 3.38TYY339 pKa = 11.32NSGGFSLPTSTTIQFPNSPFVQTYY363 pKa = 10.7LMEE366 pKa = 4.38LVWTGGAVATLLPTFTATNAVFGQAFYY393 pKa = 10.74GPPSITTTAVQVITLVVQTTPNTVPVVTLSTTLVTLPTVPTVNIQITQQPNLITGVV449 pKa = 3.36
MM1 pKa = 8.12DD2 pKa = 5.06GNGDD6 pKa = 3.83VPEE9 pKa = 4.67GWTDD13 pKa = 5.12PITMAEE19 pKa = 3.4IRR21 pKa = 11.84KK22 pKa = 9.32YY23 pKa = 10.47IKK25 pKa = 10.76NSDD28 pKa = 3.48YY29 pKa = 11.64DD30 pKa = 3.56KK31 pKa = 10.92TKK33 pKa = 10.84ARR35 pKa = 11.84AIRR38 pKa = 11.84SLVSRR43 pKa = 11.84RR44 pKa = 11.84PRR46 pKa = 11.84RR47 pKa = 11.84SYY49 pKa = 10.38RR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84SNRR56 pKa = 11.84RR57 pKa = 11.84GSRR60 pKa = 11.84RR61 pKa = 11.84RR62 pKa = 11.84GYY64 pKa = 9.91RR65 pKa = 11.84GYY67 pKa = 10.61GAYY70 pKa = 9.85SRR72 pKa = 11.84RR73 pKa = 11.84GSASGSIGQRR83 pKa = 11.84LGGYY87 pKa = 9.47LGTMLGGAAQNLLKK101 pKa = 10.12TYY103 pKa = 9.06TGLGAYY109 pKa = 7.24QVKK112 pKa = 10.3GNALMPGAVISNPNPHH128 pKa = 6.69GGQVFRR134 pKa = 11.84GSDD137 pKa = 3.48YY138 pKa = 11.33LGDD141 pKa = 3.65IFSSPVAGQFTNQSFPINAALEE163 pKa = 4.05QTFPKK168 pKa = 10.31LAQFLQNFDD177 pKa = 3.39QYY179 pKa = 11.43VIEE182 pKa = 4.09GLIFEE187 pKa = 4.77FRR189 pKa = 11.84SMSCDD194 pKa = 3.11SLNSTNTALGSIIAACNYY212 pKa = 10.03NVLQAPFASKK222 pKa = 10.71AAMEE226 pKa = 4.3EE227 pKa = 4.46YY228 pKa = 10.45EE229 pKa = 4.59GGVSQRR235 pKa = 11.84PSSNMQFFVEE245 pKa = 4.64CARR248 pKa = 11.84AQSPMDD254 pKa = 3.39VLYY257 pKa = 10.0TRR259 pKa = 11.84TGAIPANADD268 pKa = 2.9LRR270 pKa = 11.84MYY272 pKa = 11.05DD273 pKa = 3.88LGNFQIASQGLQGTSVNMGEE293 pKa = 4.37LWVHH297 pKa = 5.07YY298 pKa = 9.65QVAGLKK304 pKa = 9.35EE305 pKa = 3.95KK306 pKa = 10.3IYY308 pKa = 11.07AGFGNYY314 pKa = 8.69NSAYY318 pKa = 9.4RR319 pKa = 11.84AVGSGGNNSNPLGNTRR335 pKa = 11.84TDD337 pKa = 3.38TYY339 pKa = 11.32NSGGFSLPTSTTIQFPNSPFVQTYY363 pKa = 10.7LMEE366 pKa = 4.38LVWTGGAVATLLPTFTATNAVFGQAFYY393 pKa = 10.74GPPSITTTAVQVITLVVQTTPNTVPVVTLSTTLVTLPTVPTVNIQITQQPNLITGVV449 pKa = 3.36
Molecular weight: 48.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
828 |
379 |
449 |
414.0 |
46.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.367 ± 0.684 | 0.966 ± 0.233 |
4.469 ± 1.231 | 5.556 ± 2.257 |
3.986 ± 0.367 | 7.729 ± 2.143 |
1.449 ± 0.786 | 5.314 ± 0.499 |
4.952 ± 2.133 | 7.005 ± 0.27 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.019 ± 0.446 | 5.918 ± 0.598 |
5.797 ± 0.179 | 4.831 ± 0.751 |
5.918 ± 0.249 | 6.884 ± 0.887 |
6.763 ± 1.679 | 5.556 ± 0.881 |
1.57 ± 0.706 | 4.952 ± 0.041 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
