
Fusarium graminearum dsRNA mycovirus-4
Taxonomy: Viruses; Riboviria; dsRNA viruses; unclassified dsRNA viruses
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
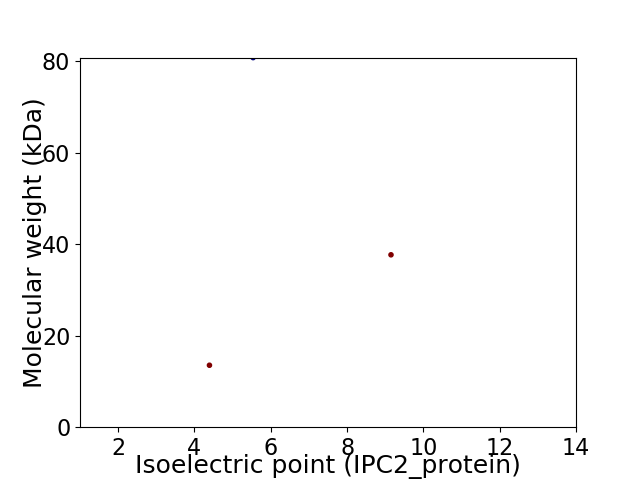
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
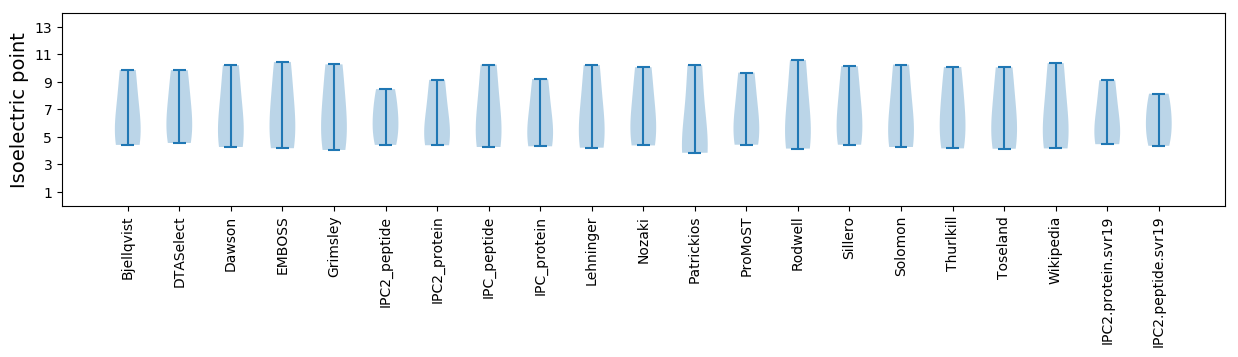
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D0TZ33|D0TZ33_9VIRU Uncharacterized protein OS=Fusarium graminearum dsRNA mycovirus-4 OX=687918 PE=4 SV=1
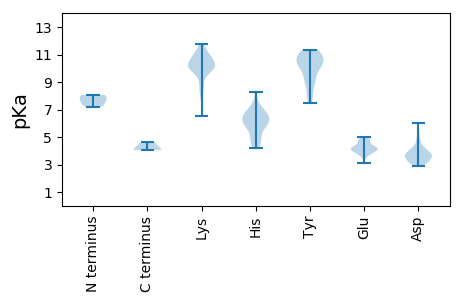
MM1 pKa = 8.06RR2 pKa = 11.84EE3 pKa = 3.87ILYY6 pKa = 10.46HH7 pKa = 4.73MWEE10 pKa = 4.26GDD12 pKa = 3.2MHH14 pKa = 6.6GLLRR18 pKa = 11.84LVTRR22 pKa = 11.84TLEE25 pKa = 4.3SEE27 pKa = 4.29AEE29 pKa = 3.97SDD31 pKa = 5.09NGDD34 pKa = 3.33EE35 pKa = 4.3EE36 pKa = 4.26QWIGSTMVPGYY47 pKa = 8.78PALPARR53 pKa = 11.84RR54 pKa = 11.84ADD56 pKa = 3.45YY57 pKa = 10.6GRR59 pKa = 11.84PFLDD63 pKa = 3.22VLRR66 pKa = 11.84WATNAAVAQGYY77 pKa = 7.63ATAQRR82 pKa = 11.84IVEE85 pKa = 4.06QDD87 pKa = 3.1PDD89 pKa = 3.17QDD91 pKa = 4.32RR92 pKa = 11.84EE93 pKa = 4.39LEE95 pKa = 4.16CLEE98 pKa = 4.29VVQDD102 pKa = 4.59AMVTGDD108 pKa = 4.11GRR110 pKa = 11.84FRR112 pKa = 11.84DD113 pKa = 3.29QWVFLL118 pKa = 4.1
MM1 pKa = 8.06RR2 pKa = 11.84EE3 pKa = 3.87ILYY6 pKa = 10.46HH7 pKa = 4.73MWEE10 pKa = 4.26GDD12 pKa = 3.2MHH14 pKa = 6.6GLLRR18 pKa = 11.84LVTRR22 pKa = 11.84TLEE25 pKa = 4.3SEE27 pKa = 4.29AEE29 pKa = 3.97SDD31 pKa = 5.09NGDD34 pKa = 3.33EE35 pKa = 4.3EE36 pKa = 4.26QWIGSTMVPGYY47 pKa = 8.78PALPARR53 pKa = 11.84RR54 pKa = 11.84ADD56 pKa = 3.45YY57 pKa = 10.6GRR59 pKa = 11.84PFLDD63 pKa = 3.22VLRR66 pKa = 11.84WATNAAVAQGYY77 pKa = 7.63ATAQRR82 pKa = 11.84IVEE85 pKa = 4.06QDD87 pKa = 3.1PDD89 pKa = 3.17QDD91 pKa = 4.32RR92 pKa = 11.84EE93 pKa = 4.39LEE95 pKa = 4.16CLEE98 pKa = 4.29VVQDD102 pKa = 4.59AMVTGDD108 pKa = 4.11GRR110 pKa = 11.84FRR112 pKa = 11.84DD113 pKa = 3.29QWVFLL118 pKa = 4.1
Molecular weight: 13.55 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D0TZ33|D0TZ33_9VIRU Uncharacterized protein OS=Fusarium graminearum dsRNA mycovirus-4 OX=687918 PE=4 SV=1
MM1 pKa = 7.74ADD3 pKa = 3.49MRR5 pKa = 11.84QMFAGATRR13 pKa = 11.84DD14 pKa = 3.42VSFNPIPGLEE24 pKa = 3.89NSYY27 pKa = 11.23LPIPAKK33 pKa = 9.94TDD35 pKa = 3.16SLTQLSKK42 pKa = 11.46KK43 pKa = 10.02MGDD46 pKa = 3.09LRR48 pKa = 11.84KK49 pKa = 9.83VVEE52 pKa = 4.7SGHH55 pKa = 5.78SLAVATGMRR64 pKa = 11.84GAAFDD69 pKa = 3.87TVNPEE74 pKa = 3.7EE75 pKa = 4.23KK76 pKa = 10.3AIAEE80 pKa = 4.04MGVAEE85 pKa = 4.34KK86 pKa = 10.39EE87 pKa = 4.16QYY89 pKa = 10.42LAWKK93 pKa = 9.98GGARR97 pKa = 11.84LSDD100 pKa = 4.19FDD102 pKa = 4.78PATCHH107 pKa = 6.33EE108 pKa = 4.84PTSKK112 pKa = 10.2WNGSIATLDD121 pKa = 2.88KK122 pKa = 10.26WLRR125 pKa = 11.84GLRR128 pKa = 11.84AMYY131 pKa = 10.51ADD133 pKa = 4.26EE134 pKa = 4.11EE135 pKa = 4.46LQRR138 pKa = 11.84EE139 pKa = 4.28HH140 pKa = 6.87AVWFAVRR147 pKa = 11.84APFMVPTITAHH158 pKa = 5.92VKK160 pKa = 10.18VLTAGKK166 pKa = 10.87SMVDD170 pKa = 2.86QHH172 pKa = 6.95AKK174 pKa = 10.17LRR176 pKa = 11.84PEE178 pKa = 4.12RR179 pKa = 11.84MAEE182 pKa = 3.85YY183 pKa = 10.29QSAKK187 pKa = 10.32RR188 pKa = 11.84ILADD192 pKa = 2.9AAYY195 pKa = 9.0RR196 pKa = 11.84VQVIKK201 pKa = 10.97RR202 pKa = 11.84EE203 pKa = 4.3INKK206 pKa = 8.24LTQEE210 pKa = 3.7IGRR213 pKa = 11.84AEE215 pKa = 4.28DD216 pKa = 3.51VVLTRR221 pKa = 11.84VRR223 pKa = 11.84HH224 pKa = 4.46YY225 pKa = 8.04QQKK228 pKa = 10.55VGTKK232 pKa = 6.52EE233 pKa = 3.95HH234 pKa = 7.02KK235 pKa = 8.99EE236 pKa = 3.78APEE239 pKa = 3.46NRR241 pKa = 11.84NRR243 pKa = 11.84KK244 pKa = 8.31RR245 pKa = 11.84RR246 pKa = 11.84GMPALGAPTDD256 pKa = 3.98LGGMTGTGRR265 pKa = 11.84EE266 pKa = 3.87ARR268 pKa = 11.84RR269 pKa = 11.84RR270 pKa = 11.84RR271 pKa = 11.84IDD273 pKa = 3.85LGPQLGTSPYY283 pKa = 10.93SLRR286 pKa = 11.84PWLGSDD292 pKa = 3.85HH293 pKa = 7.53PSTTHH298 pKa = 6.86LVPPLSPPSMRR309 pKa = 11.84RR310 pKa = 11.84IWAISSRR317 pKa = 11.84TPQCPPAGGSRR328 pKa = 11.84SRR330 pKa = 11.84PDD332 pKa = 3.12CRR334 pKa = 11.84HH335 pKa = 5.65WEE337 pKa = 4.02LL338 pKa = 4.61
MM1 pKa = 7.74ADD3 pKa = 3.49MRR5 pKa = 11.84QMFAGATRR13 pKa = 11.84DD14 pKa = 3.42VSFNPIPGLEE24 pKa = 3.89NSYY27 pKa = 11.23LPIPAKK33 pKa = 9.94TDD35 pKa = 3.16SLTQLSKK42 pKa = 11.46KK43 pKa = 10.02MGDD46 pKa = 3.09LRR48 pKa = 11.84KK49 pKa = 9.83VVEE52 pKa = 4.7SGHH55 pKa = 5.78SLAVATGMRR64 pKa = 11.84GAAFDD69 pKa = 3.87TVNPEE74 pKa = 3.7EE75 pKa = 4.23KK76 pKa = 10.3AIAEE80 pKa = 4.04MGVAEE85 pKa = 4.34KK86 pKa = 10.39EE87 pKa = 4.16QYY89 pKa = 10.42LAWKK93 pKa = 9.98GGARR97 pKa = 11.84LSDD100 pKa = 4.19FDD102 pKa = 4.78PATCHH107 pKa = 6.33EE108 pKa = 4.84PTSKK112 pKa = 10.2WNGSIATLDD121 pKa = 2.88KK122 pKa = 10.26WLRR125 pKa = 11.84GLRR128 pKa = 11.84AMYY131 pKa = 10.51ADD133 pKa = 4.26EE134 pKa = 4.11EE135 pKa = 4.46LQRR138 pKa = 11.84EE139 pKa = 4.28HH140 pKa = 6.87AVWFAVRR147 pKa = 11.84APFMVPTITAHH158 pKa = 5.92VKK160 pKa = 10.18VLTAGKK166 pKa = 10.87SMVDD170 pKa = 2.86QHH172 pKa = 6.95AKK174 pKa = 10.17LRR176 pKa = 11.84PEE178 pKa = 4.12RR179 pKa = 11.84MAEE182 pKa = 3.85YY183 pKa = 10.29QSAKK187 pKa = 10.32RR188 pKa = 11.84ILADD192 pKa = 2.9AAYY195 pKa = 9.0RR196 pKa = 11.84VQVIKK201 pKa = 10.97RR202 pKa = 11.84EE203 pKa = 4.3INKK206 pKa = 8.24LTQEE210 pKa = 3.7IGRR213 pKa = 11.84AEE215 pKa = 4.28DD216 pKa = 3.51VVLTRR221 pKa = 11.84VRR223 pKa = 11.84HH224 pKa = 4.46YY225 pKa = 8.04QQKK228 pKa = 10.55VGTKK232 pKa = 6.52EE233 pKa = 3.95HH234 pKa = 7.02KK235 pKa = 8.99EE236 pKa = 3.78APEE239 pKa = 3.46NRR241 pKa = 11.84NRR243 pKa = 11.84KK244 pKa = 8.31RR245 pKa = 11.84RR246 pKa = 11.84GMPALGAPTDD256 pKa = 3.98LGGMTGTGRR265 pKa = 11.84EE266 pKa = 3.87ARR268 pKa = 11.84RR269 pKa = 11.84RR270 pKa = 11.84RR271 pKa = 11.84IDD273 pKa = 3.85LGPQLGTSPYY283 pKa = 10.93SLRR286 pKa = 11.84PWLGSDD292 pKa = 3.85HH293 pKa = 7.53PSTTHH298 pKa = 6.86LVPPLSPPSMRR309 pKa = 11.84RR310 pKa = 11.84IWAISSRR317 pKa = 11.84TPQCPPAGGSRR328 pKa = 11.84SRR330 pKa = 11.84PDD332 pKa = 3.12CRR334 pKa = 11.84HH335 pKa = 5.65WEE337 pKa = 4.02LL338 pKa = 4.61
Molecular weight: 37.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1168 |
118 |
712 |
389.3 |
44.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.503 ± 0.334 | 1.113 ± 0.137 |
7.277 ± 0.986 | 6.079 ± 0.915 |
3.596 ± 0.896 | 6.507 ± 0.558 |
2.911 ± 0.328 | 3.339 ± 0.204 |
4.11 ± 1.072 | 9.075 ± 0.546 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.911 ± 0.62 | 2.397 ± 0.273 |
6.079 ± 0.589 | 4.366 ± 0.438 |
8.305 ± 0.748 | 5.736 ± 0.828 |
5.908 ± 0.215 | 5.479 ± 0.616 |
2.055 ± 0.37 | 3.253 ± 0.473 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
